Flare™
Ligand alignment
Compare congeneric and diverse active ligands to understand SAR and build models and pharmacophores
Get insight into how your series compares with other known inhibitors
Using electrostatic and shape properties of ligands enables the comparison of diverse molecules that bind at the same active site giving you insight into how your series compares with other known inhibitors. Our field based alignment and scoring method is the engine behind successful large scale virtual screening campaigns in Blaze™ and scaffold hopping in Spark™. Use it in Flare to compare ligands from different series, enabling SAR transfer, design bespoke scaffolds, or conduct small virtual screens either in the GUI or through the command line pyflare executable.
Generate reliable alignments of multiple ligands
Cresset’s patented ligand comparison method, which aligns and scores molecules, generates biologically relevant comparisons of active ligands; and has a proven track record in virtual screening, scaffold hopping and pharmacophore determination.
Alternatively, use substructure alignment to generate inputs into QSAR model building or FEP calculations. Flexible options and extensive ways to influence the results using known SAR give results that you can trust with minimal human intervention.
Right: Chemical analogues aligned to the crystallographic ligand of 6RLN at the active site, using Cresset field points.
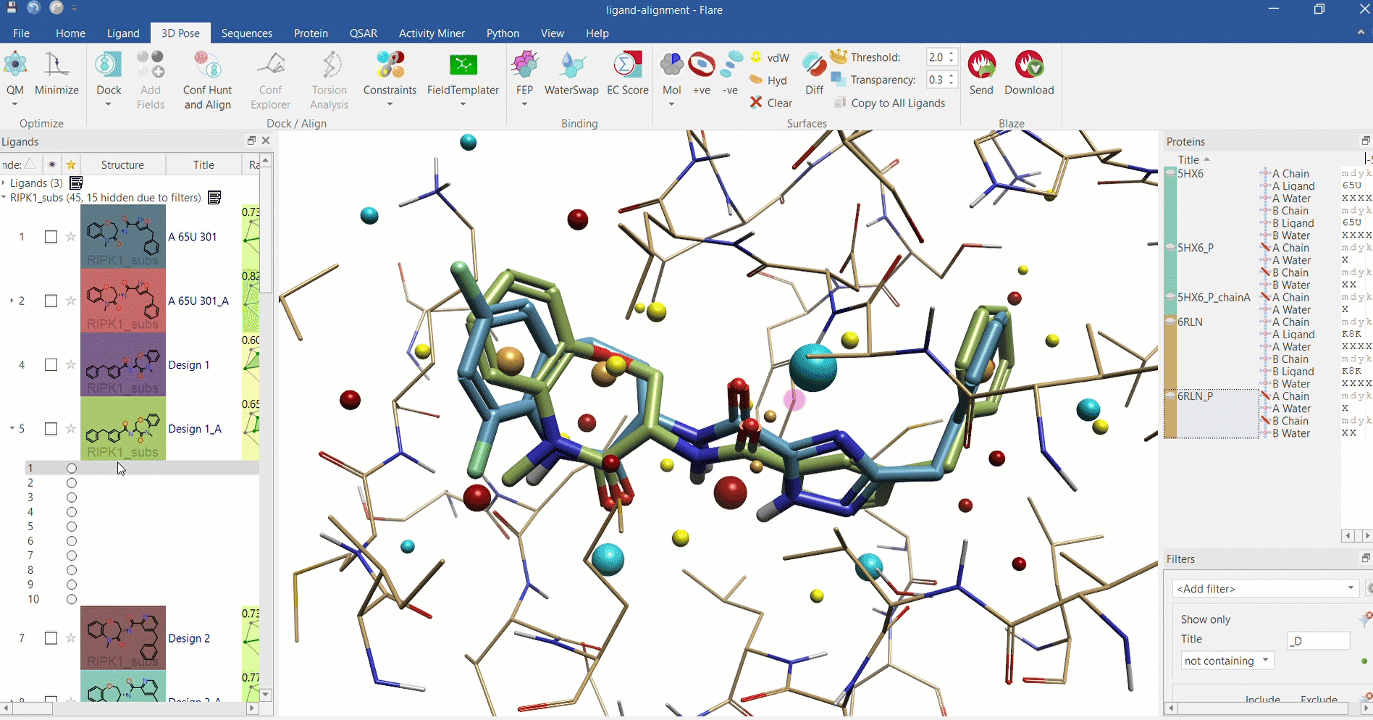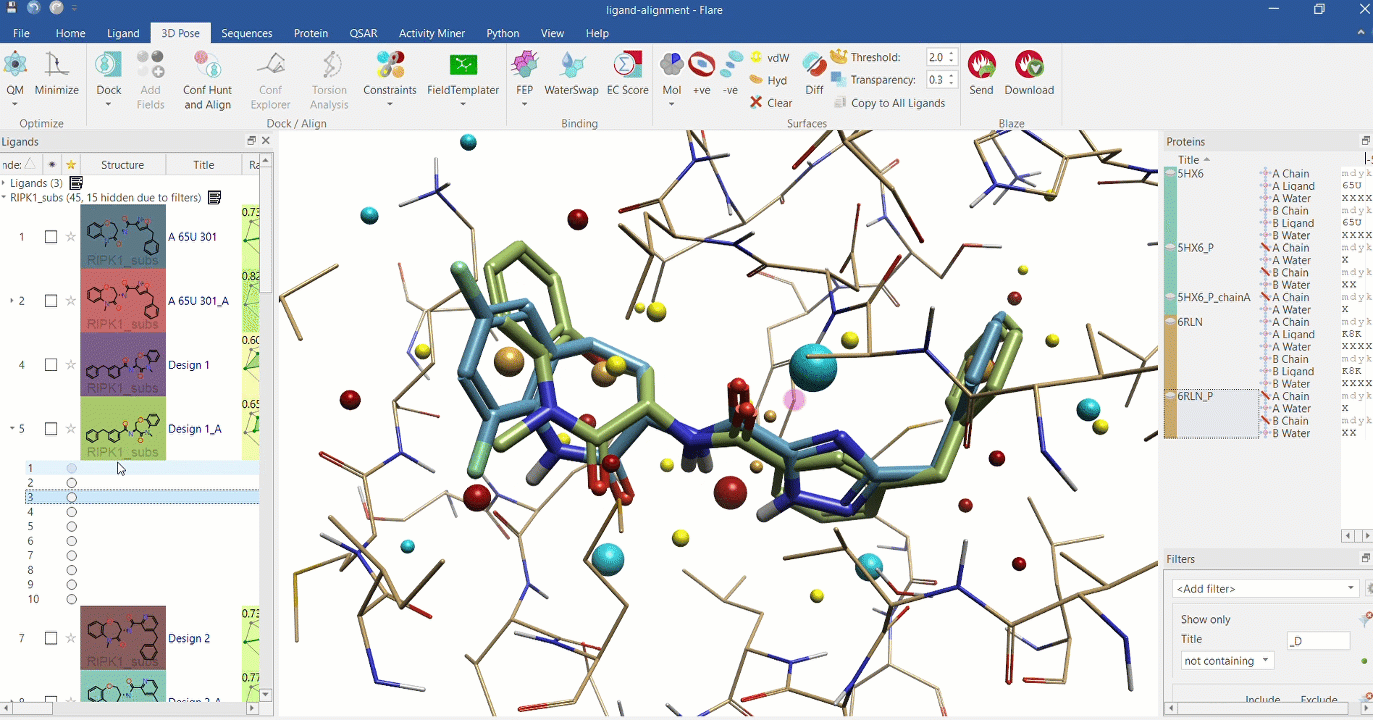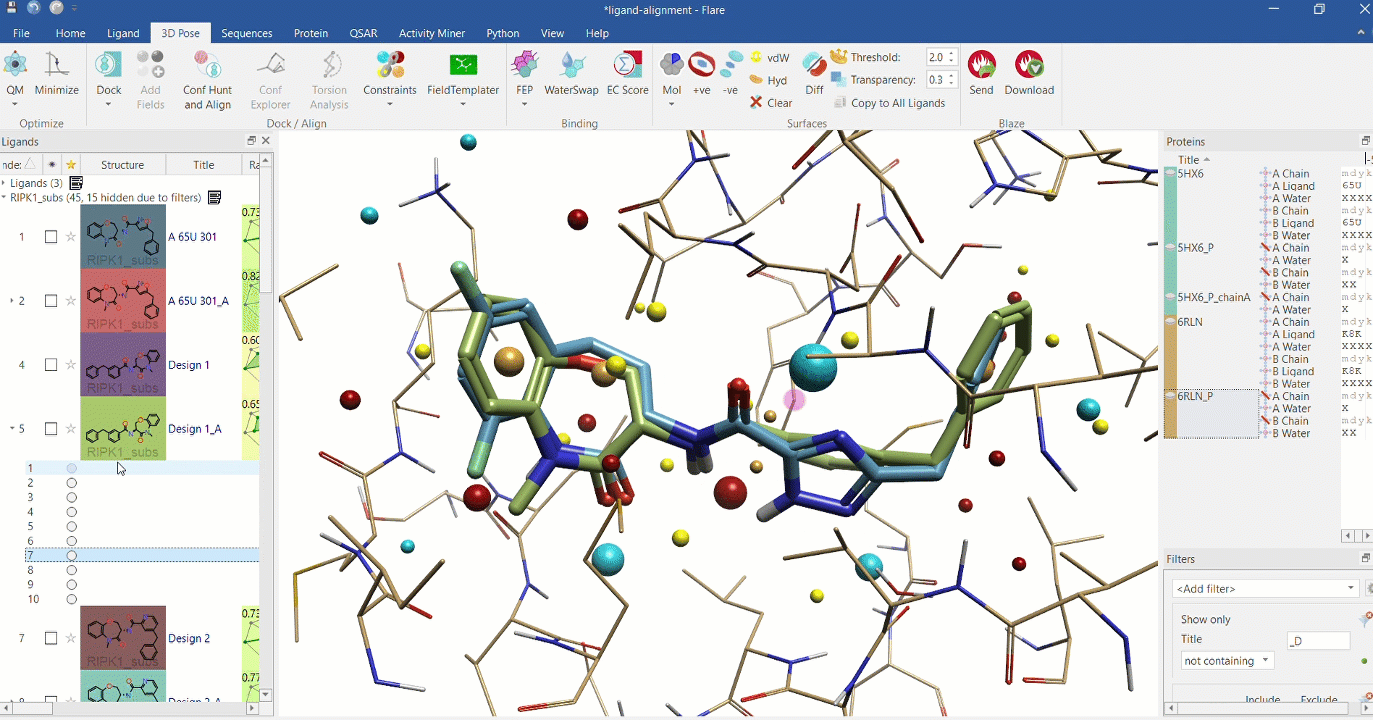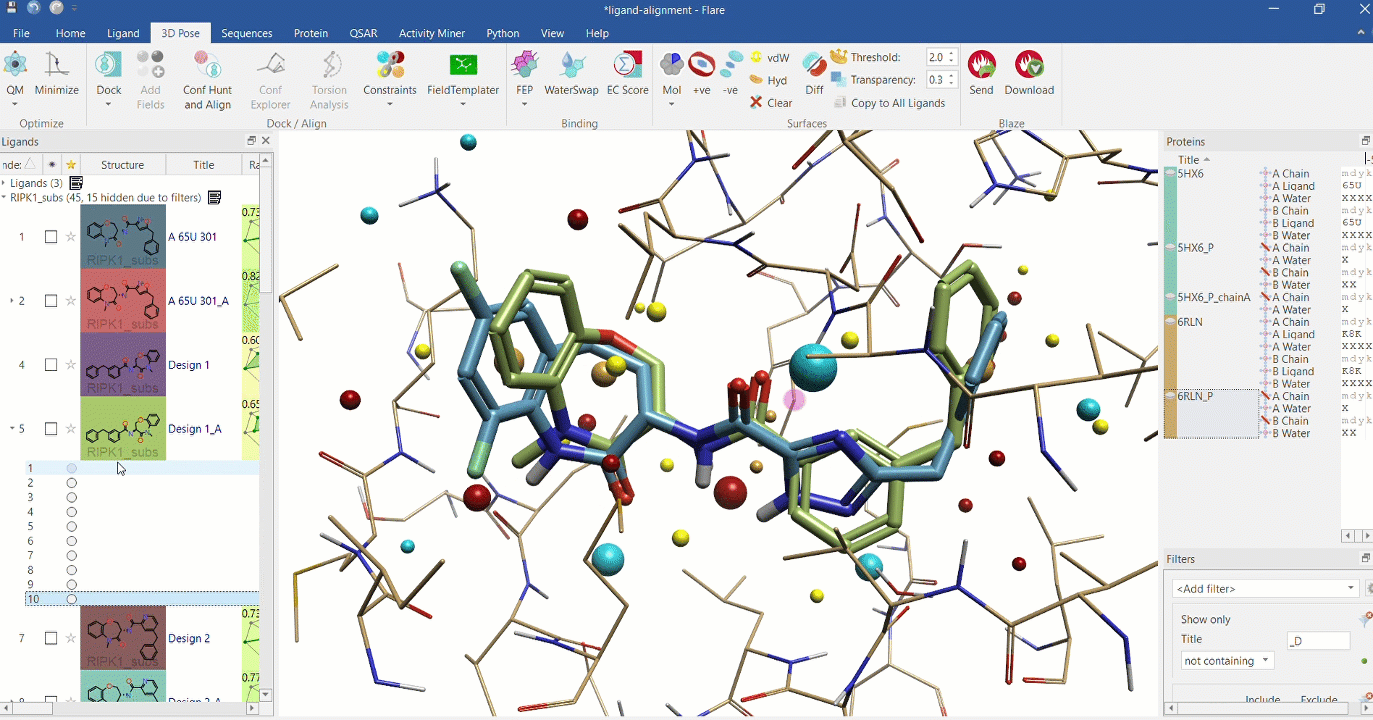
Substructure alignment
The extensive substructure alignment methods in Flare enable you to generate inputs to SAR investigations.
- Align all your molecules to a single chemotype as input to QSAR or FEP experiments
- Automatically align to multiple reference structures in a single experiment
- Control the alignment through substructure matching rules and SMARTS patterns
Perfect alignment using existing SAR
Whether aligning ligands using electrostatics or substructure, existing SAR will dictate your view of the results.
Flare provides multiple ways to bias the alignment towards the existing knowledge that you have on the project.
- Choose to use field constraints to enforce alignments based on electrostatics
- Choose pharmacophore constraints to enforce a particular functionality to make an interaction
- Add a protein structure to use as an excluded volume in the alignments