flare™
Flare MM/GBSA
Rapidly interrogate the binding affinity of your ligands through binding free energy prediction
Molecular Mechanics, General Born surface area (MM/GBSA) is a method to calculate the ligand-protein binding free energy. The MM/GBSA binding free energy is derived from molecular mechanics: it offers the advantage of being more theoretically rigorous compared to the empirical scoring functions used by molecular docking, and the benefit of being far less computationally expensive than relative binding free energy simulations.
- Run MM/GBSA experiments to evaluate the binding free energy of ligands from a single conformation in a fraction of the time and cost compared to relative binding free energy calculations
- Score hundreds of compounds from the GUI, thousands from the command line
- Options to minimize the ligand and protein, yielding improved accuracy in your binding free energy predictions
- A wide variety of different implicit solvent models are available, making MM/GBSA adaptable to your protein-ligand system of interest
- Use MM/GBSA for a high-throughput assessment of ligand binding affinities within your compound series at the end of a docking experiment, prioritizing your molecule designs with more confidence
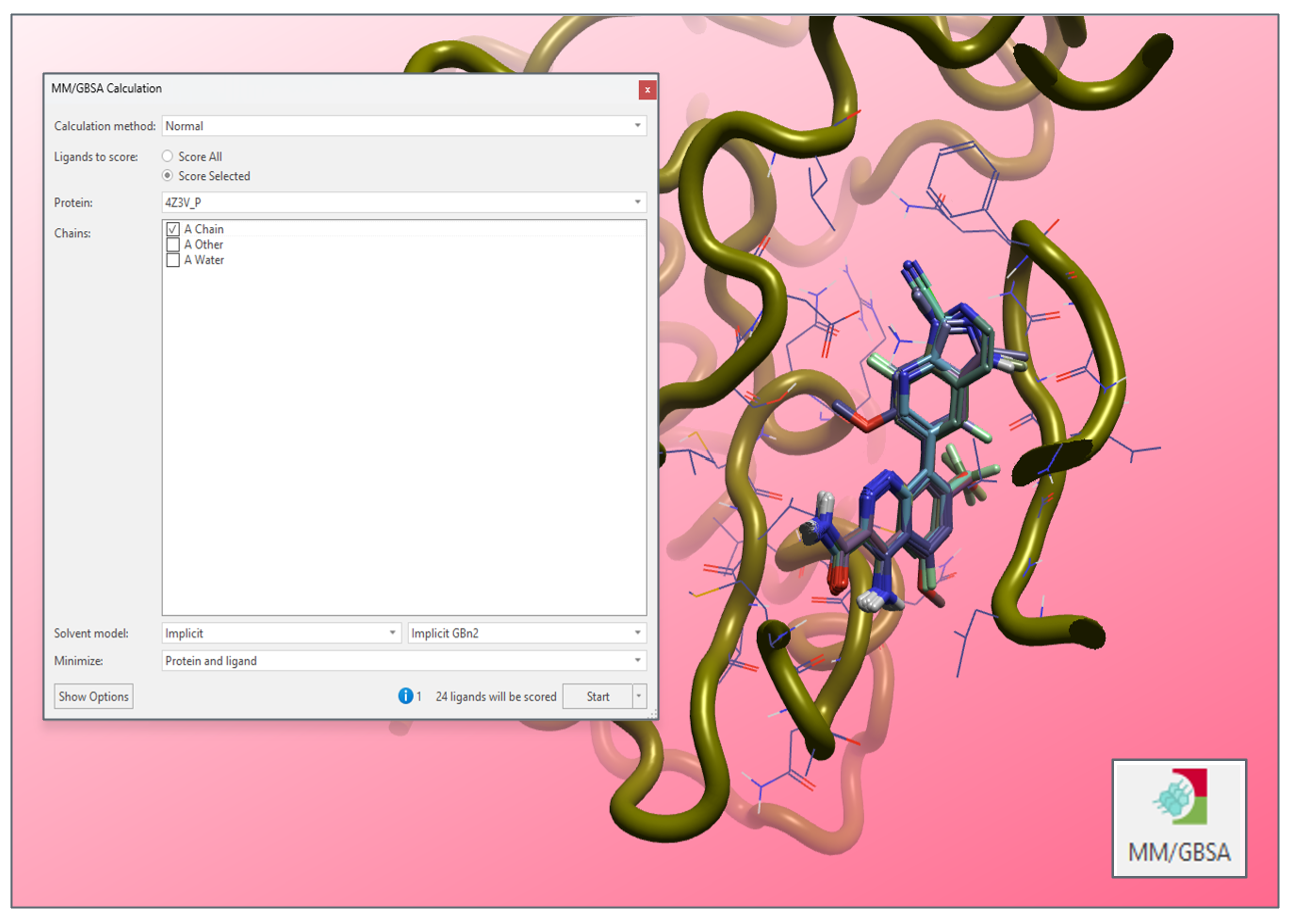
MM/GBSA provides a means to quickly interrogate the binding affinity of bound ligand conformations, through computationally efficient free energy measurements
References and acknowledgements
S. Genheden, U. Ryde, The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities, Expert Opin Drug Discov. 2015,10, 5
Flare
Explore further to see how Flare can add fresh insights into your structure-based design
Explore further