Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Spark is the most advanced and complex bioisostere finding tool available today. While it excels at finding bioisosteric replacements that match the steric, pharmacophoric and electronic nature of the original molecule, we realize that the output of a Spark search is only the beginning. Spark suggests new molecules, so someone needs to make them. Given the time and effort that goes into even a simple synthesis, we need to make sure that Spark’s suggestions are as robust as they can be to maximize the changes of success for each Spark result.
Spark’s internal algorithms do a lot of complicated work to ensure that the result molecules are chemically sensible. When you are stitching a fragment into a new molecule there are many subtle effects that you have to take account of:
On this last point, Spark carries out a rotation around the newly-formed bond in order to estimate the amount of strain energy carried by that bond in the assigned torsion angle. For speed, this rotational scan is performed holding the parts of the molecule on each side of the bond rigid, and as a result the computed strain energy is only an estimate. However, even this estimated strain energy can be very useful to flag up cases where the new molecule is in an energetically unfavorable conformation and hence would need further investigation before it could be recommended for synthesis.
While this purely computational procedure works well, it would be nice to bring in some more empirical knowledge gleaned from the vast numbers of small molecule crystal structures that are available in the Cambridge Structural Database (CSD). Luckily, this analysis has already been done by Prof. Rarey’s group at the University of Hamburg, resulting in a pair of papers detailing a hierarchical set of rules to determine preferred values for torsion angles in small drug-like molecules.1, 2 This rule set, called the Torsion Library, has been incorporated into the most recent release of Spark (version 10.4).
Whenever a new product molecule is formed, Spark applies the Torsion Library rules to the newly-formed bonds, and highlights cases where the torsion angle is not one that is frequently observed in the CSD. This doesn’t automatically exclude that result from consideration (there may be a reason such as steric clashes for the uncommon torsion), but it does flag that result molecule as needing careful inspection. The Torsion Library results are automatically displayed for all results and can be used in Spark’s filters like any other result column.
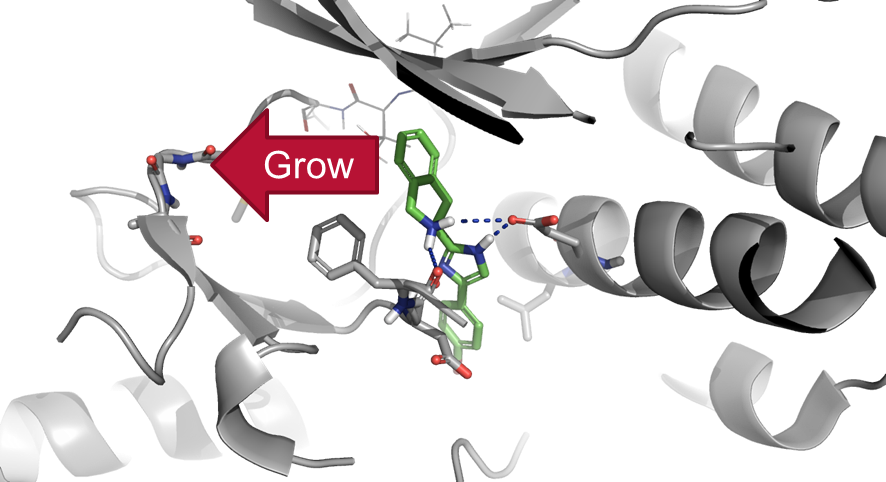
As a quick example of the usefulness of the Torsion Library, we have re-examined the 2011 case study FieldStere V3.0 Example 2: Fragment Growing. In this study we were performing a fragment growing experiment in p38, using an existing ligand to guide the growth of a fragment towards the hinge binding region (Figure 1). If we had particular chemistries in mind, then instead of searching the general reagent databases, we could search specific reagent databases to find what commercially-available reagents we could use.
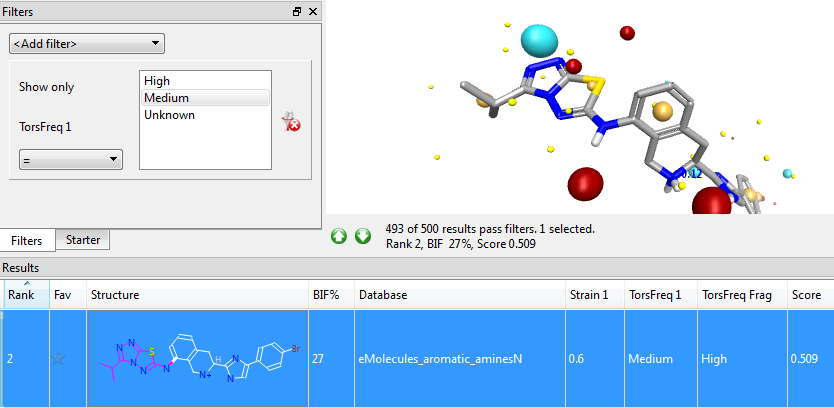
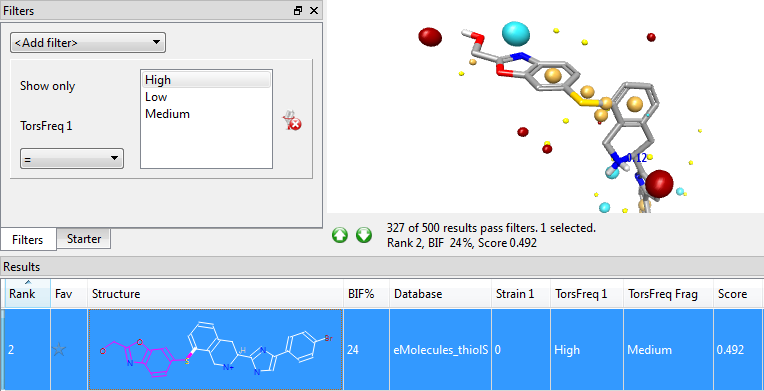
Adding the Torsion Library to Spark makes the results even more robust and useful, allowing you to see at a glance what the known experimental conformational preferences of small molecules say about the conformer quality in your Spark results. The new feature is available now as part of the Spark 10.4 release – try a free evaluation today.
1 Shärfer et al., J. Med. Chem. 2013, 56(5), 2016
2 Guba et al., JCIM 2016, 56, 1