Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Version 8 of Flare, Cresset’s CADD workbench, brings new and enhanced scientific features and methods, including the new Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) method for calculating the binding free energy of ligand-protein complexes, a significant expansion of Flare FEP domain of applicability, Grand Canonical Nonequilibrium Candidate Monte Carlo (GCNCMC) for GIST water analysis, calculation and display of HOMO/LUMO orbitals for QM calculations, more than 100 new reactions for Library Enumeration, new and enhanced features for protein and homology modeling.
In this release we have also further expanded the choice of highly visual tools to troubleshoot Flare FEP experiments and analyze dynamics trajectories. Tools for creating ligand custom parameters for dynamics and FEP simulations and preparing ligand structures for further studies have also been further developed and enhanced.
Furthermore, new Flare Profiles can be applied to align the Flare GUI to your current licensing level, making the platform more efficient and simpler to use.
MM/GBSA1 is a method to calculate the binding free energy for ligand-protein complexes, and provides a good balance between accuracy and computational efficiency. As it uses molecular mechanics, it is more theoretically rigorous than the empirical scoring functions used, for example, by molecular docking, giving more accurate results in a variety of systems (Figure 1). At the same time, it is less computationally expensive than relative binding free energy simulations such as those in Flare FEP, making it possible to rapidly score hundreds of compounds directly from the Flare GUI, and thousands from the command line.
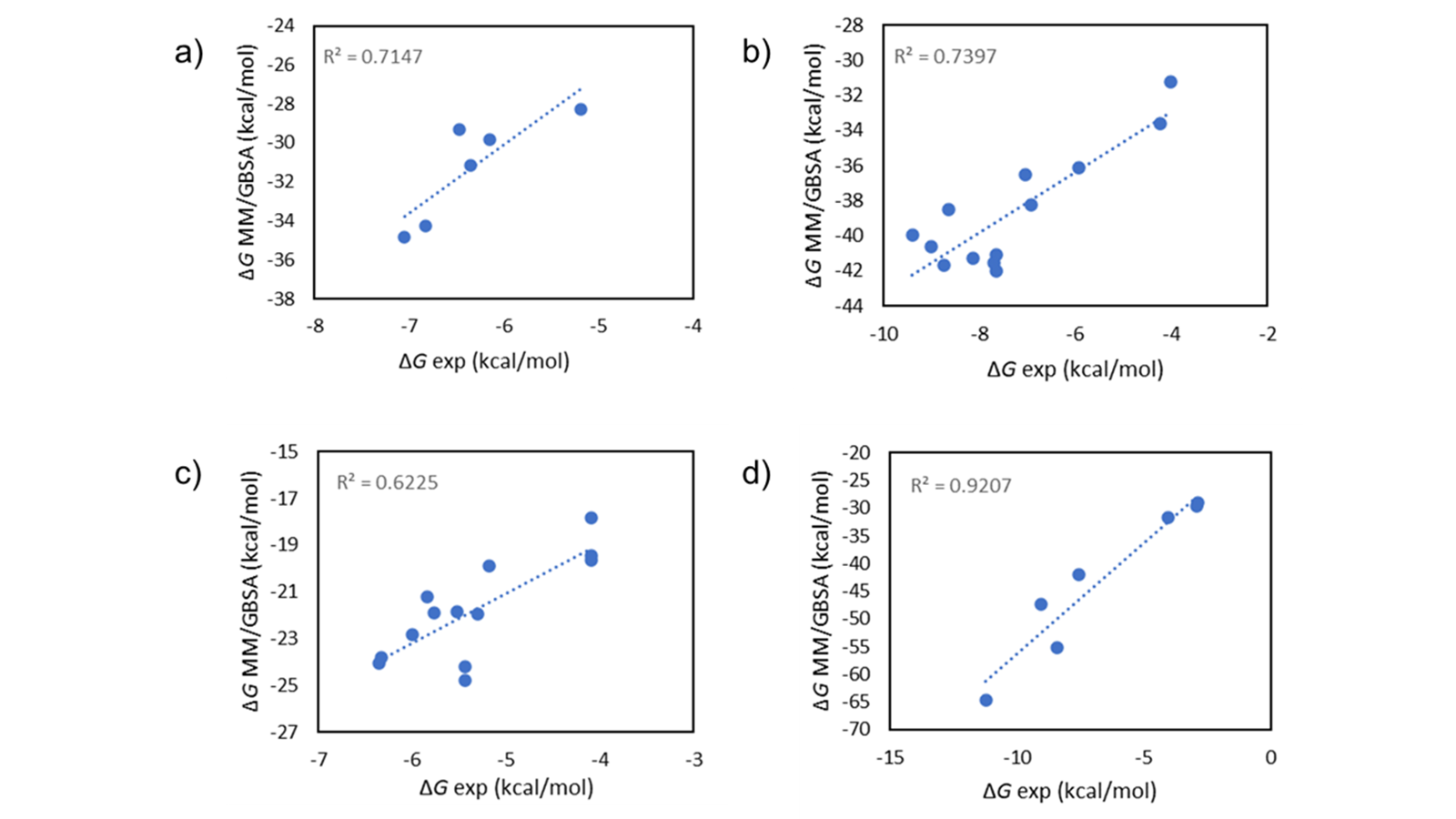
Figure 1. Experimental vs. calculated ∆G values on a FBDD dataset2 using Flare’s implementation of MM/GBSA single-point scoring
For these reasons, MM/GBSA can be used for a high-throughput assessment of binding affinities for congeneric ligands, for example after a Flare docking or Spark™ R-group replacement experiment, enabling you to confidently prioritize your molecule designs.
Flare’s flexible implementation of MM/GBSA (Figure 2) gives the choice of using different versions of OpenFF as well as the AMBER GAFF/GAFF2 force fields to parametrize the ligands. A variety of different implicit solvent models are available, making MM/GBSA adaptable to your protein-ligand system of interest, while options to minimize the ligand and protein to reduce elastic strain and potential electrostatic clashes within your protein-ligand system yield improved accuracy to the binding free energy predictions.
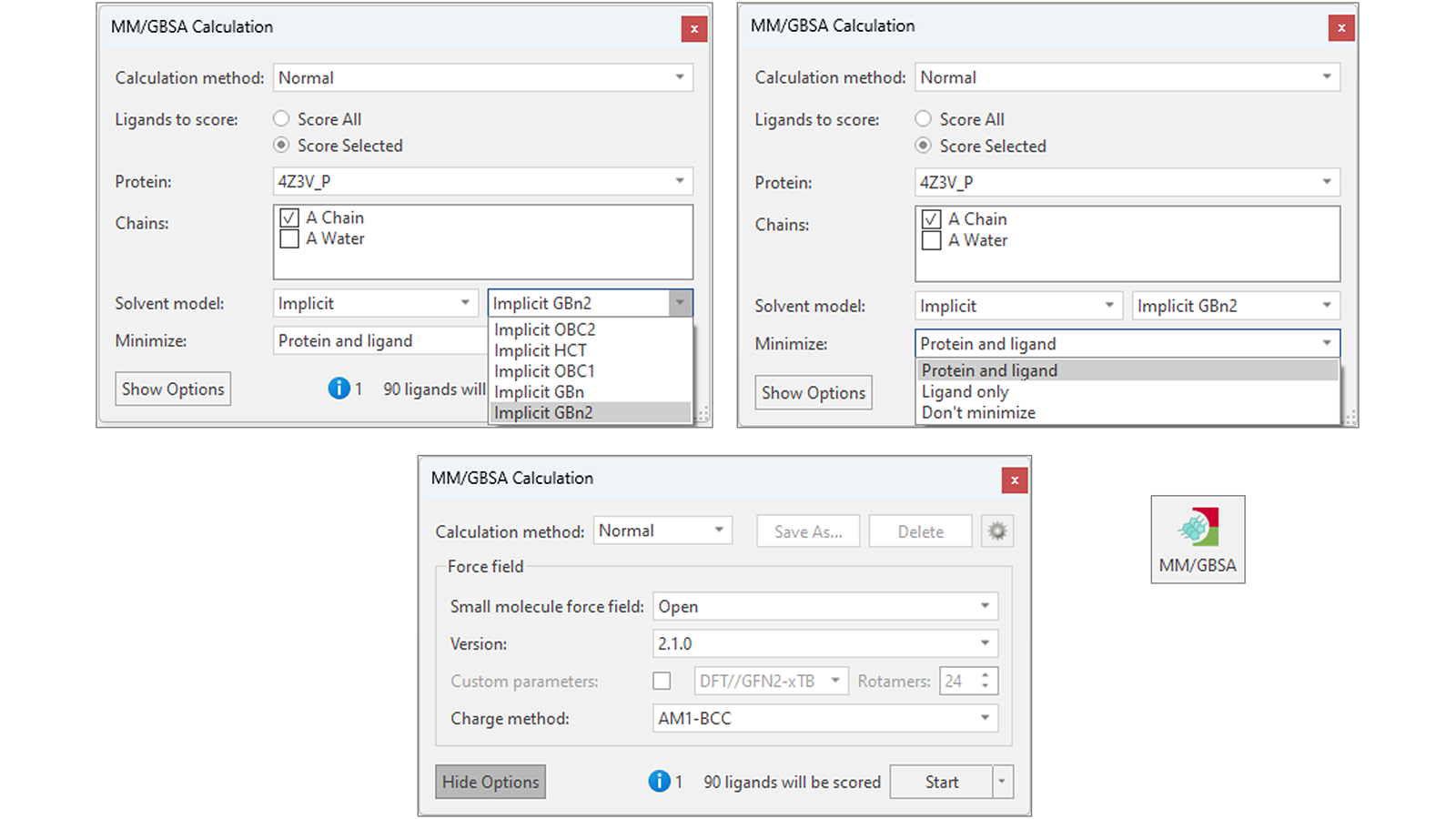
Figure 2. MM/GBSA’s flexible implementation in Flare offers different implicit solvent models (top left), choices for minimizing the ligands and target protein (top right), and different small molecule force fields for parametrizing the ligands to be scored (bottom).
Flare FEP in this new release of Flare supports projects including ligands with different net charge, significantly expanding the applicability of Flare FEP to even more real-life drug discovery projects.
Predictive performance is in line with data reported in the literature for these networks: an example of which is shown in Figure 3.
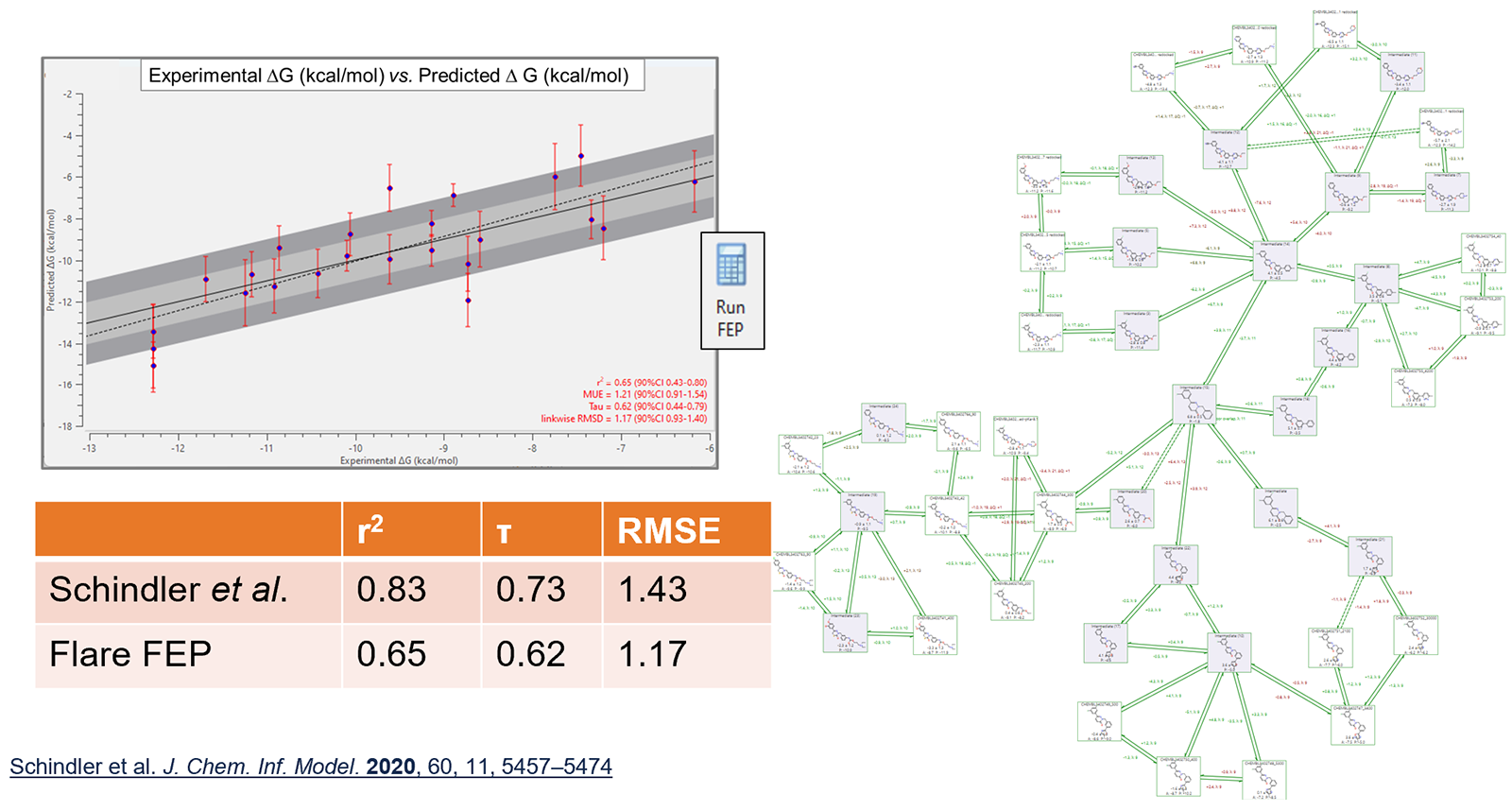
Figure 3. Predictive performance of Flare FEP on a c-Met data set, compared to that reported in the Schindler et al. paper.3
This release is also equipped with additional, highly visual tools to understand and troubleshoot the results of Flare FEP calculations.
The Torsion Plot (Figure 4) shows a histogram of the torsion angles adopted by each rotatable bond of the ligands involved in a certain Flare FEP transformation, computed for each frame of the transformation in both the free and bound state. This plot is useful to visualize possibly unexpected conformational changes which occur between the start and end molecules: potentially a sign that the system is under sampled and may need to be re-run for a longer simulation duration.
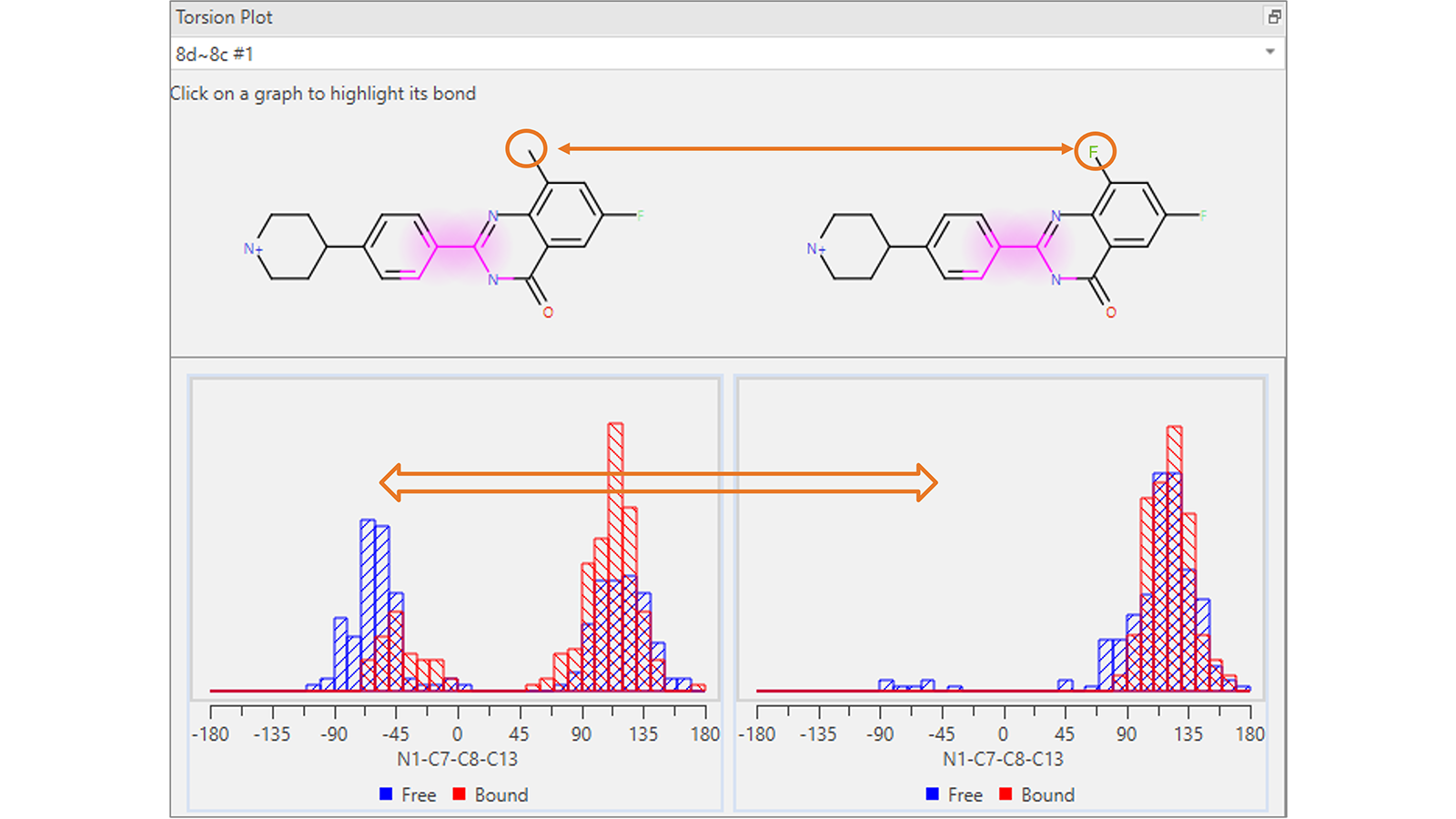
Figure 4. This Torsion Plot shows an unexpected difference in the distribution of torsion angles for the highlighted rotatable bond in the common scaffold of the molecules involved in the Flare FEP transformation.
The Convergence Plot is useful to assess the reliability and convergence of FEP simulations over time for each transformation in the Flare FEP project. By plotting the ∆G and associated error at each fraction of the simulation time (forward line) and for the corresponding last section of the trajectory (reverse line), it provides a highly effective visual tool to identify transformations with sub-optimal convergence, as in the example shown in Figure 5. In optimally converged transformations, the forward and reverse lines approach each other well before the right-hand side of the plot: this does not happen for the transformation shown in Figure 5, which may require a longer simulation.
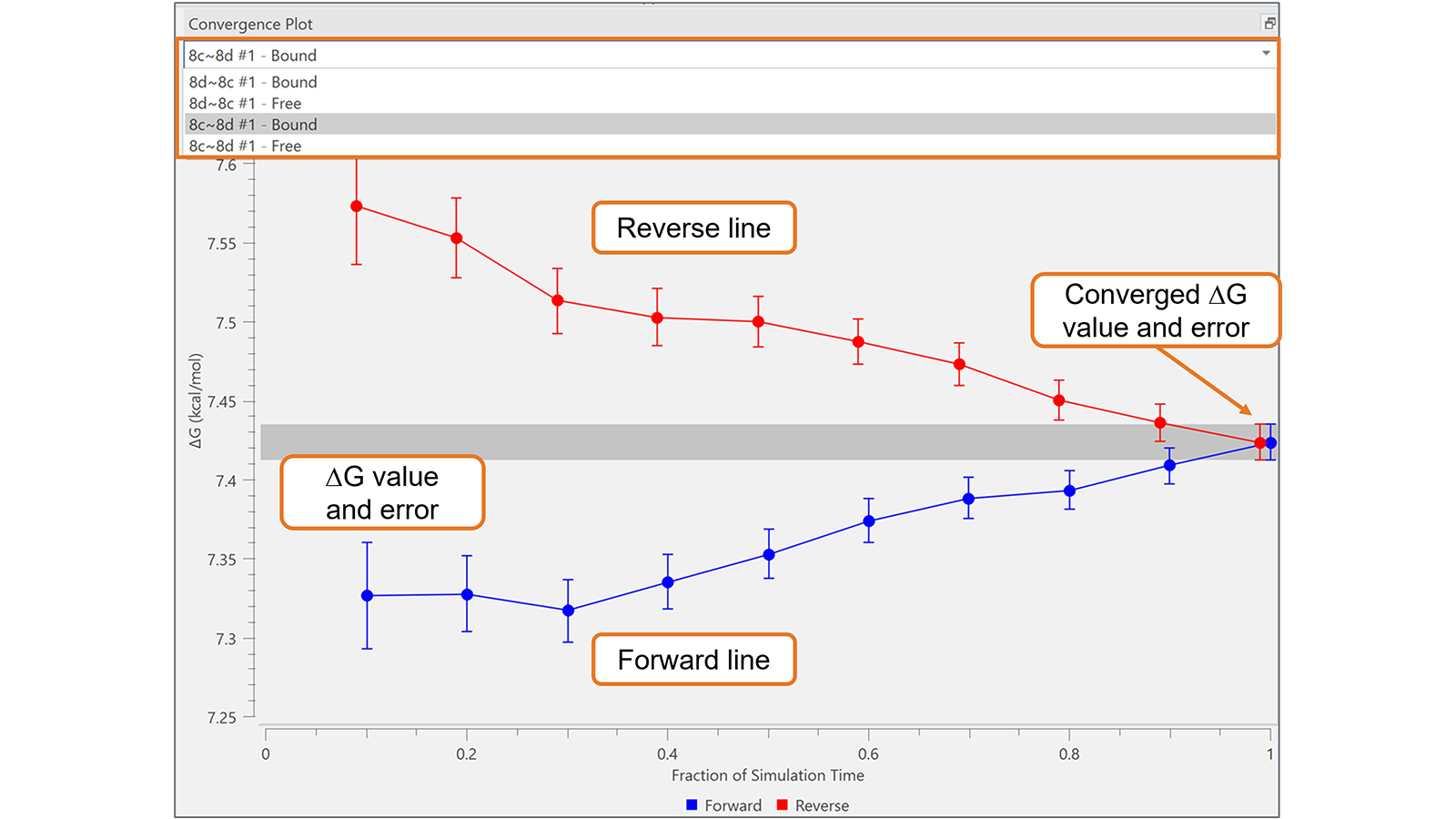
Figure 5. The Convergence Plot is useful to monitor transformation with sub-optimal convergence, as in this example, which may require a longer simulation.
Finally, the new Contacts table (Figure 6) is useful to monitor the contacts formed between the ligand being transformed and its neighboring molecules (protein, water molecules, co-factors, etc.) during each lambda window of the transformation. Contacts which change unexpectedly are highlighted, making it easier to identify noteworthy contact behavior.
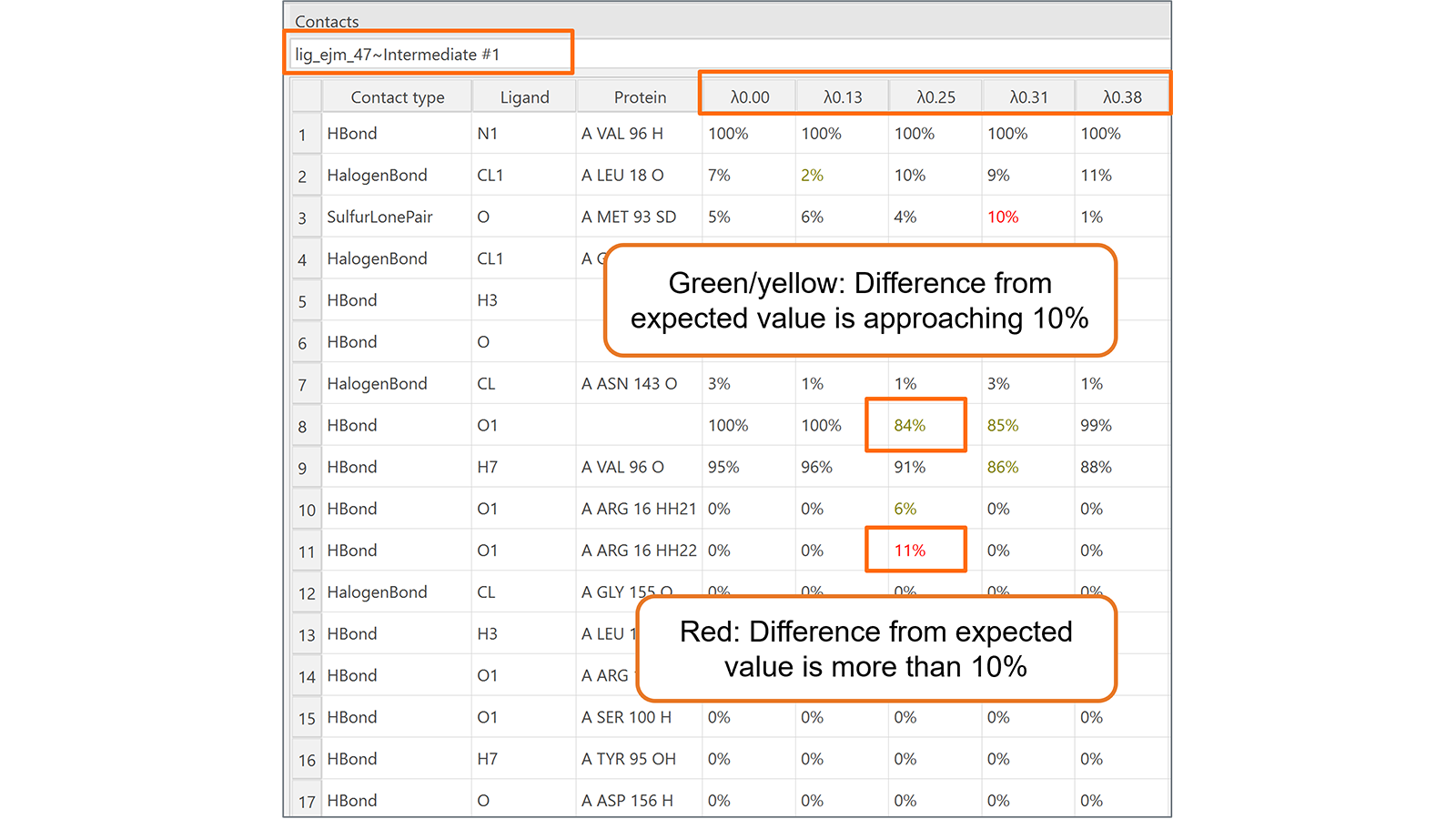
Figure 6. The new Contacts table for Flare FEP calculations monitors the contacts formed between the ligand being transformed and its neighboring molecules (protein, water molecules, co-factors, etc.) during each lambda window of the transformation.
We have further expanded the implementation of Quantum Mechanics calculations for small molecules in Flare to include the capability to compute and display HOMO (Highest Occupied Molecular Orbital) and LUMO (Lowest Unoccupied Molecular Orbital) orbitals. This is very useful, together with the calculation of the HOMO/LUMO gap, to gain insights into the reactivity and metabolic behavior of small molecules.
For example (Figure 7 – top) it is known from literature4 that the nucleophilic substitution of 2,4-dichloro-5-methylpyrimidine with piperidine happens preferentially at the 4 position, rather than at the 2 position. The computation and visualization of the LUMO for 2,4-dichloro-5-methylpyrimidine explains the experimental behavior. C2 and C5 have no associated LUMO lobes, while both C4 and C6 are associated with the largest lobes: the nucleophilic substitution is accordingly favored at one of these positions.
In a second example5 (Figure 7 – bottom), inspecting the HOMO for naphthalene explains while the electrophilic attack of nitric acid takes place preferably in the C1 (rather than C2) position, as this is associated with a larger HOMO lobe, indicating that this position is more electron-rich.
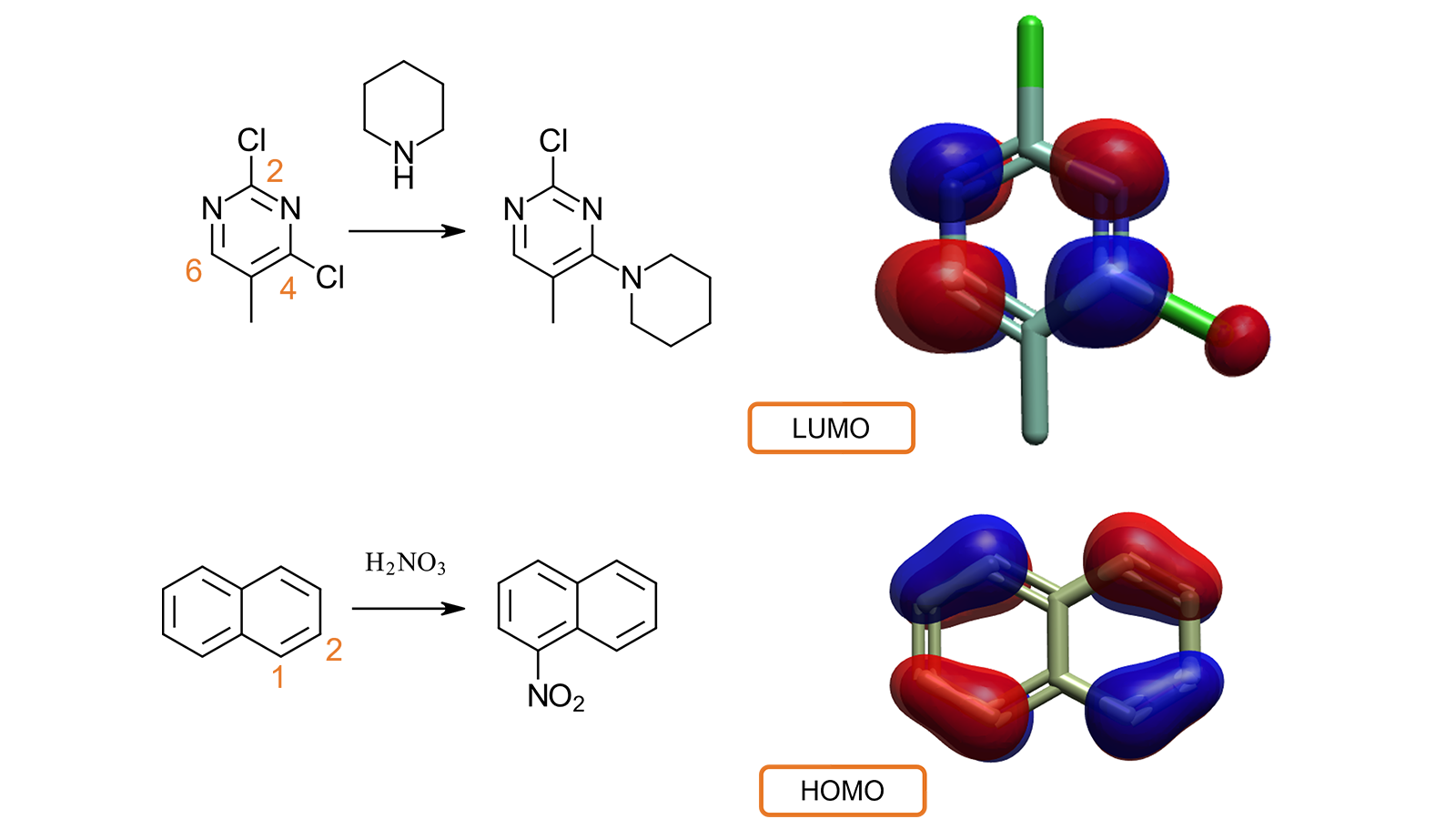
Figure 7. Visualizing LUMO and HOMO helps understanding of the reactivity and metabolic behavior of small molecules.
We have significantly expanded Flare’s collection of pre-made enumeration reactions, which now features more than 150 examples. These widely used synthetic chemistry reactions cover a large variety of transformations, including the formation of bicyclic/monocyclic aromatic rings and non-aromatic rings, C-C and N|O|S-C bond formation, deprotection, oxidation and reduction reactions.
A new 'search reactions' feature has also been added to quickly find the desired reaction (Figure 8).
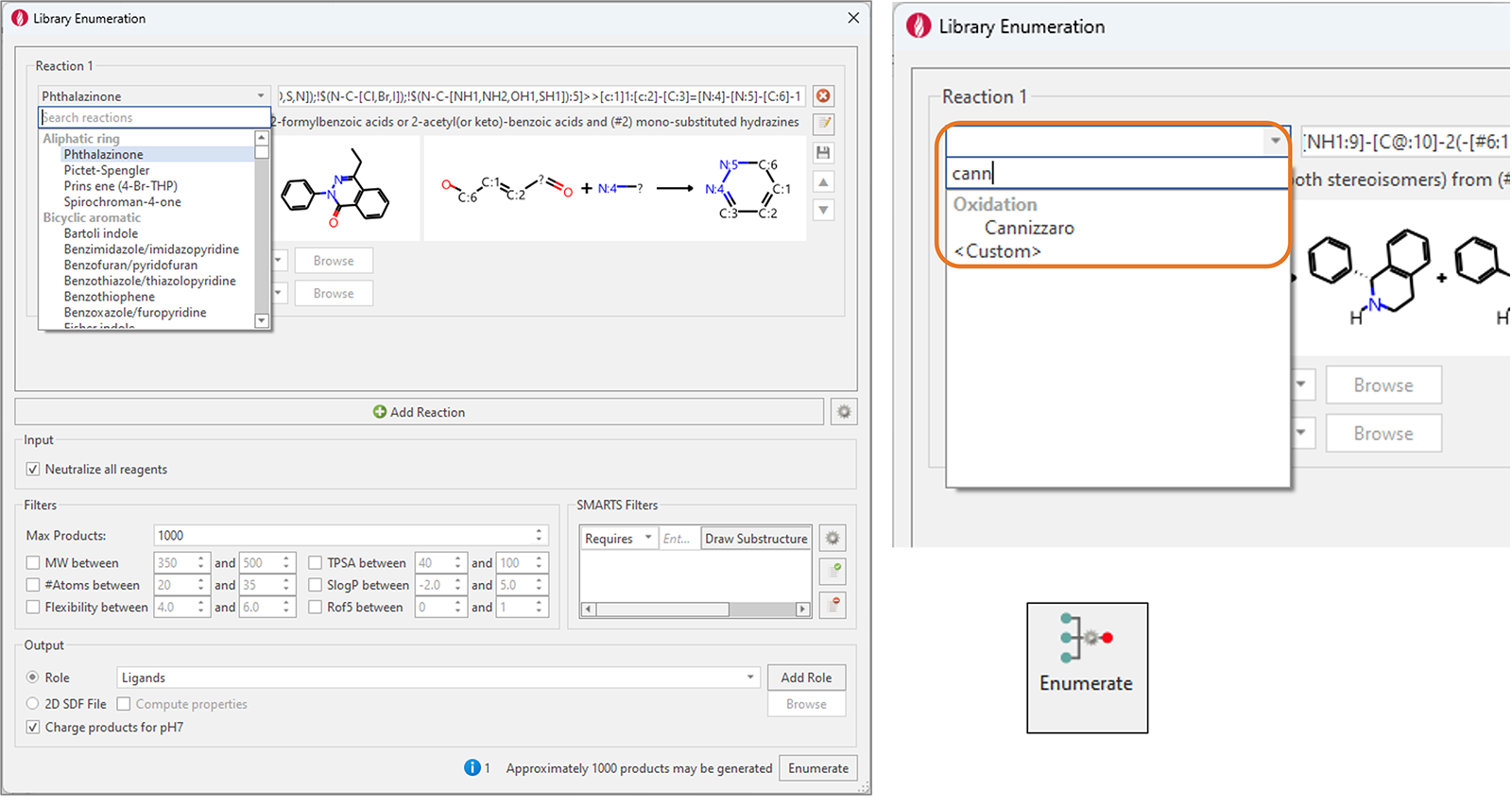
Figure 8. Left: Library Enumeration in Flare V8 features more than 100 new reactions covering a variety of widely used synthetic chemistry reactions. Right: the new ‘search reactions’ feature makes it easy to find the desired reaction.
Robust ligand preparation is a necessary and important step to prepare your small molecules for further molecular modeling studies, fixing their protonation and tautomeric state, stripping away any salts, and enumerating (when necessary) undefined stereocenters.
In Flare V8 we have added a new ‘Pop to 3D and minimize’ option (Figure 9) to complete your ligand preparation with the generation of sensible 3D structures for your ligands, ready for further calculations such as docking and scoring or ligand-based alignment.
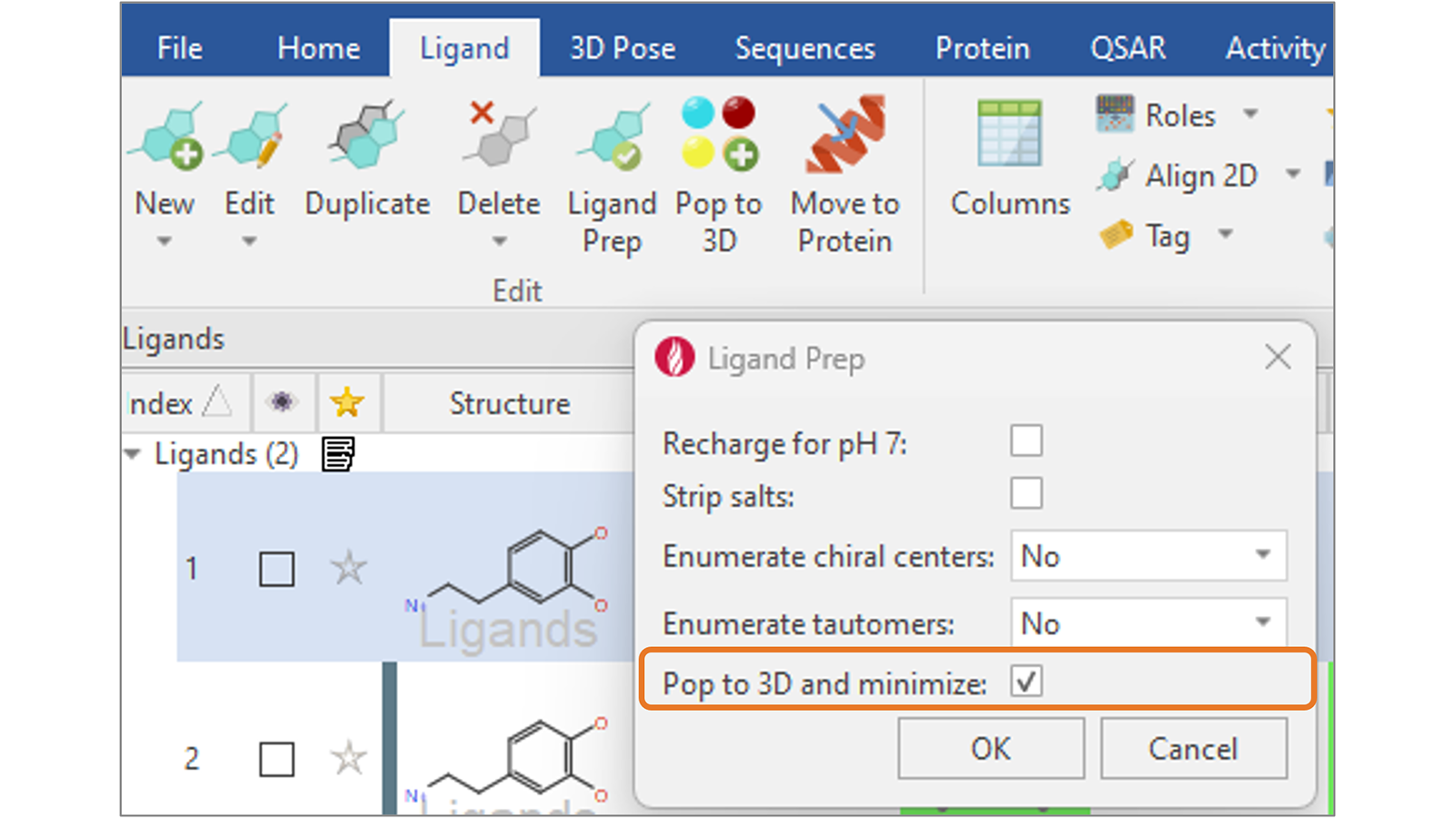
Figure 9. Use the new 'Pop to 3D and minimize' option to complete your ligand preparation with the generation of sensible 3D structures for your ligands.
The new 'Radius of Gyration' (RG) plot (Figure 10) adds to the growing collection of highly visual tools available in Flare to analyze the results of Molecular Dynamics studies.
This plot shows the RG value for the alpha carbons of the protein as a function of time and can be used to characterize the compactness or size of a biomolecule, typically a protein or a peptide. Smaller RG values indicate a more compact or tightly folded structure, while larger RG values suggest a more extended or disordered conformation. Accordingly, RG plots can be used to study the structural changes, flexibility, and dynamics of biomolecules, which can be crucial in understanding their function and behavior.
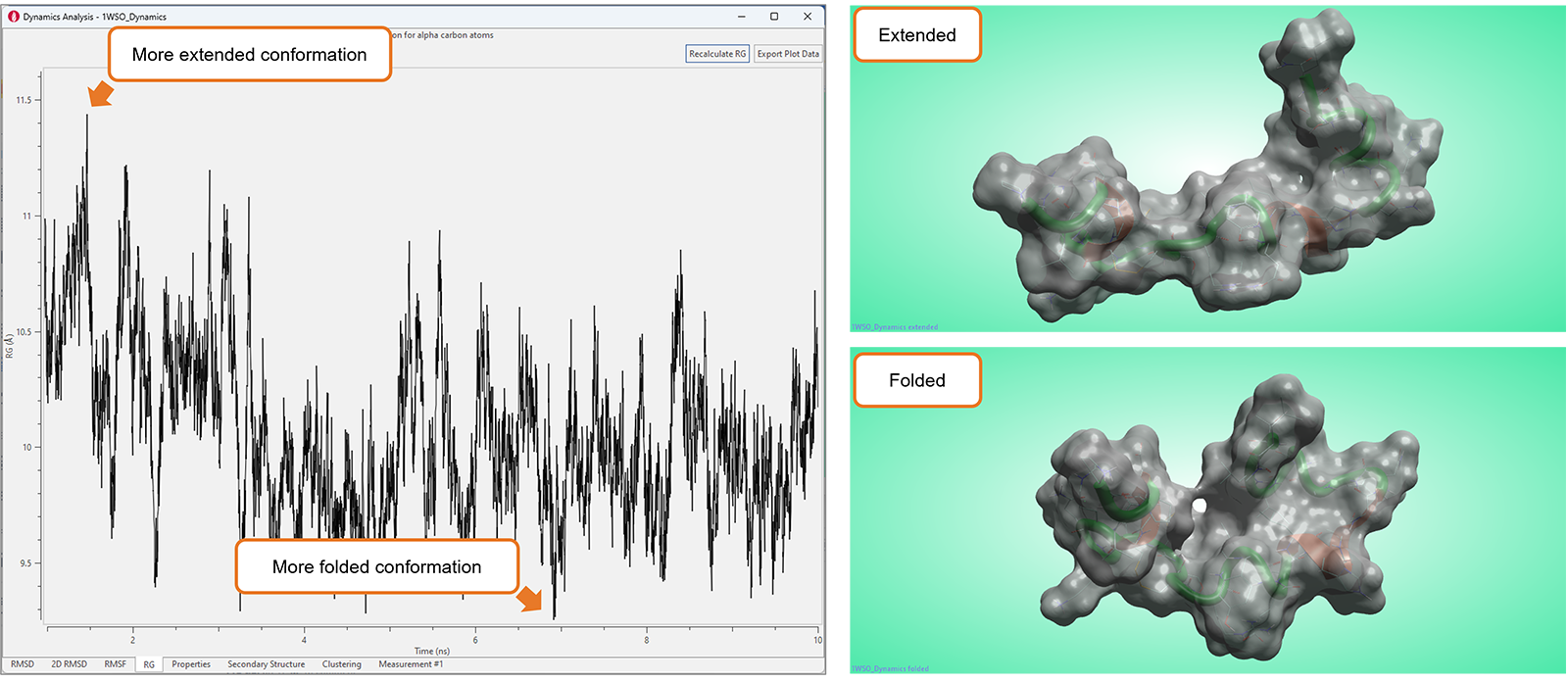
Figure 10. Significant fluctuations in the RG plot typically map conformational changes in the biomolecule under study.
In Flare V8 we have also further enhanced the Contacts table, reporting contact statistics for favorable ligand-protein interactions observed during the simulation, to include ligand interactions with water molecules (Figure 11).
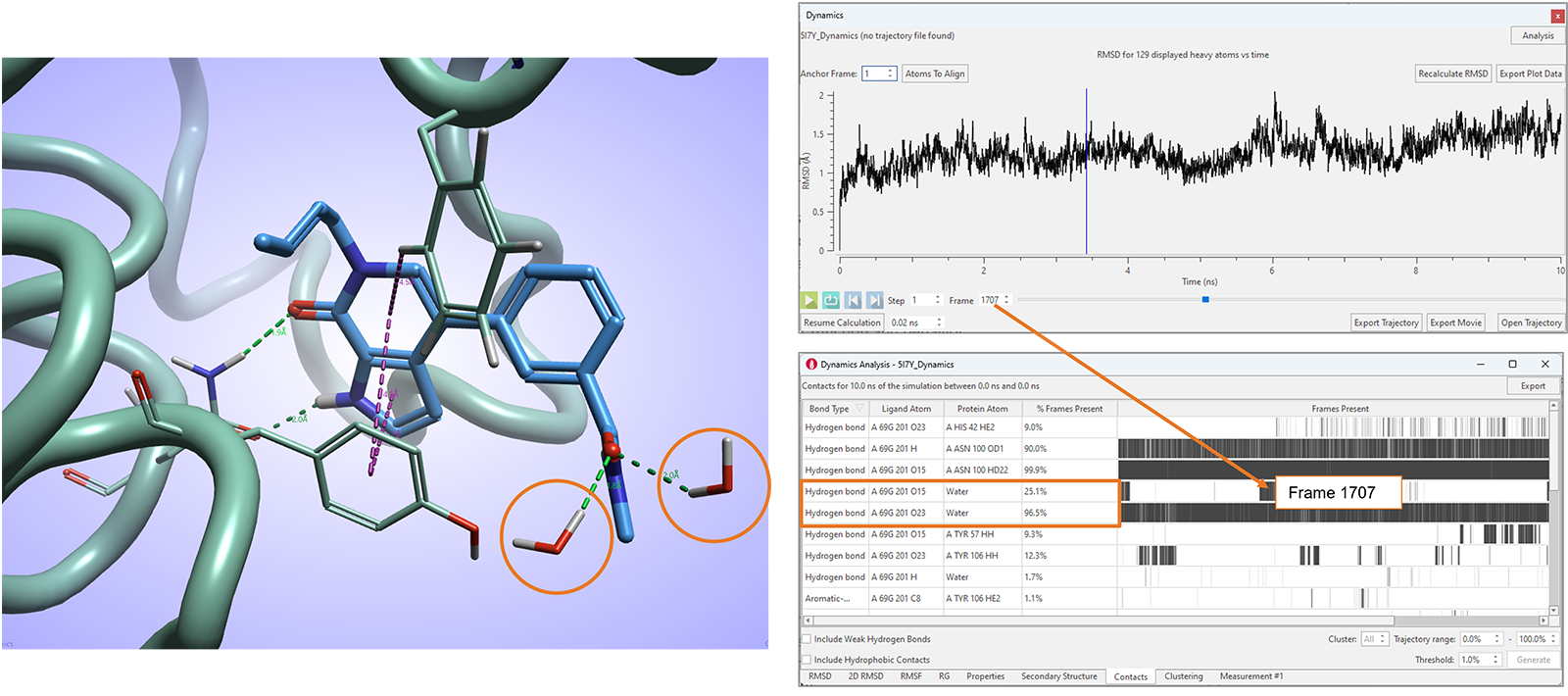
Figure 11. The Contacts analysis table in Flare V8 maps favorable ligand-protein and ligand-water interactions.
Further enhancements for Molecular Dynamics in Flare include the capability to use Grand Canonical Nonequilibrium Candidate Monte Carlo (GCNCMC)6,7 also on un-liganded, apo active sites and to export non-consecutive frames of a Dynamics trajectory.
Generation of accurate OpenFF torsion parameters for your ligands in Flare V8 is simplified by a dedicated ‘Custom Parameters’ function. With one or multiple ligands selected, pressing this button shows a widget giving you the option to choose the OpenFF version and the method to use to generate custom parameters (DFT//GFN2-xTB, GFN2-xTB or ANI-2X), as well as the number of rotamers to use for the calculation (Figure 12 top-right).
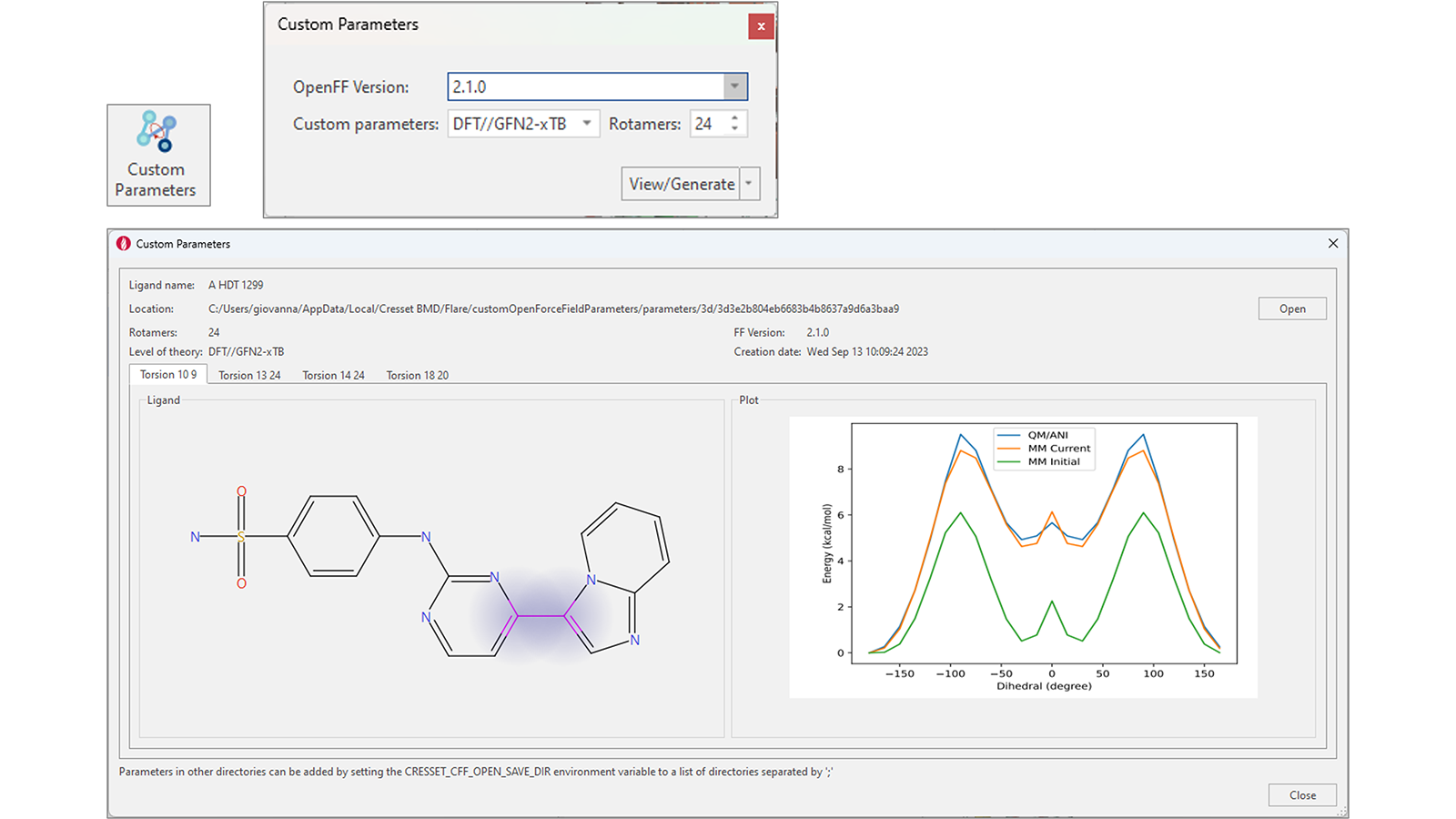
Figure 12. An enhanced workflow for the generation of accurate torsion force field parameters.
At the end of the calculation, the custom parameters can be stored locally, or in a shared location, and will be used automatically by subsequent Dynamics and Flare FEP calculations.
If custom parameters already exist, pressing the 'View/Generate' button will instead open a window showing the results of the calculation for each rotatable bond in ligand of interest (Figure 12 – bottom).
This release of Flare includes new and enhanced tools to facilitate the modeling of protein structures.
The ‘New Conformation’ function8 in the ‘Protein’ tab can be used to explore alternative, sensible conformations for the side chain of a selected residue. You can explore one side chain conformation at a time by repeatedly pressing the button or create all the conformations which avoid steric clashes with the rest of the protein by choosing the corresponding option, which will save them as alternate conformations (Figure 13).
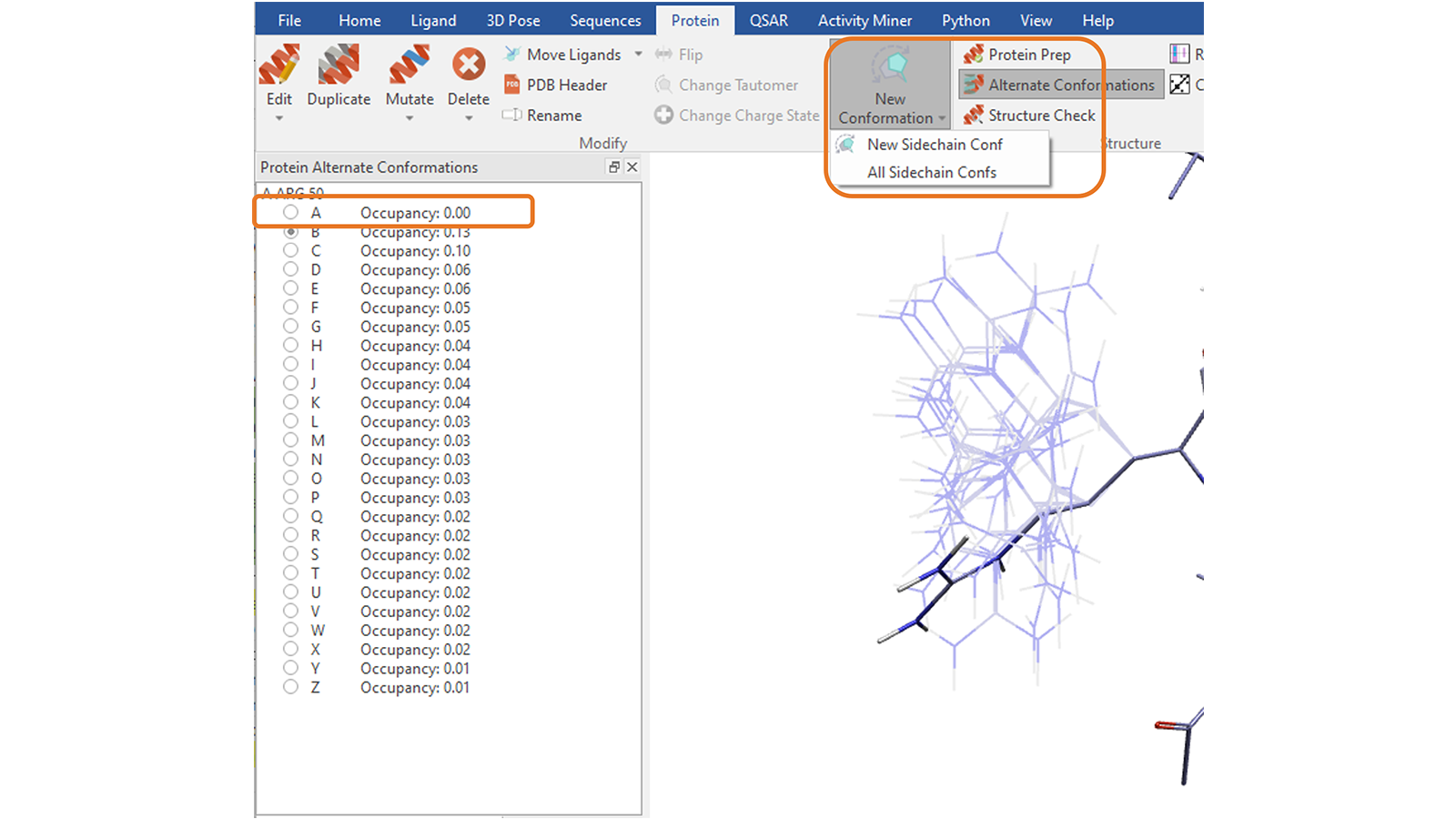
Figure 13. Create one conformation at a time, or all sensible conformations, for the side chain of a picked protein residue. Choosing to create all sidechains conformations will save them as alternate conformations, with the first entry in the list being the original conformation.
New functions to facilitate protein and homology modeling have been added to the ‘Sequences’ tab (Figure 14), including:
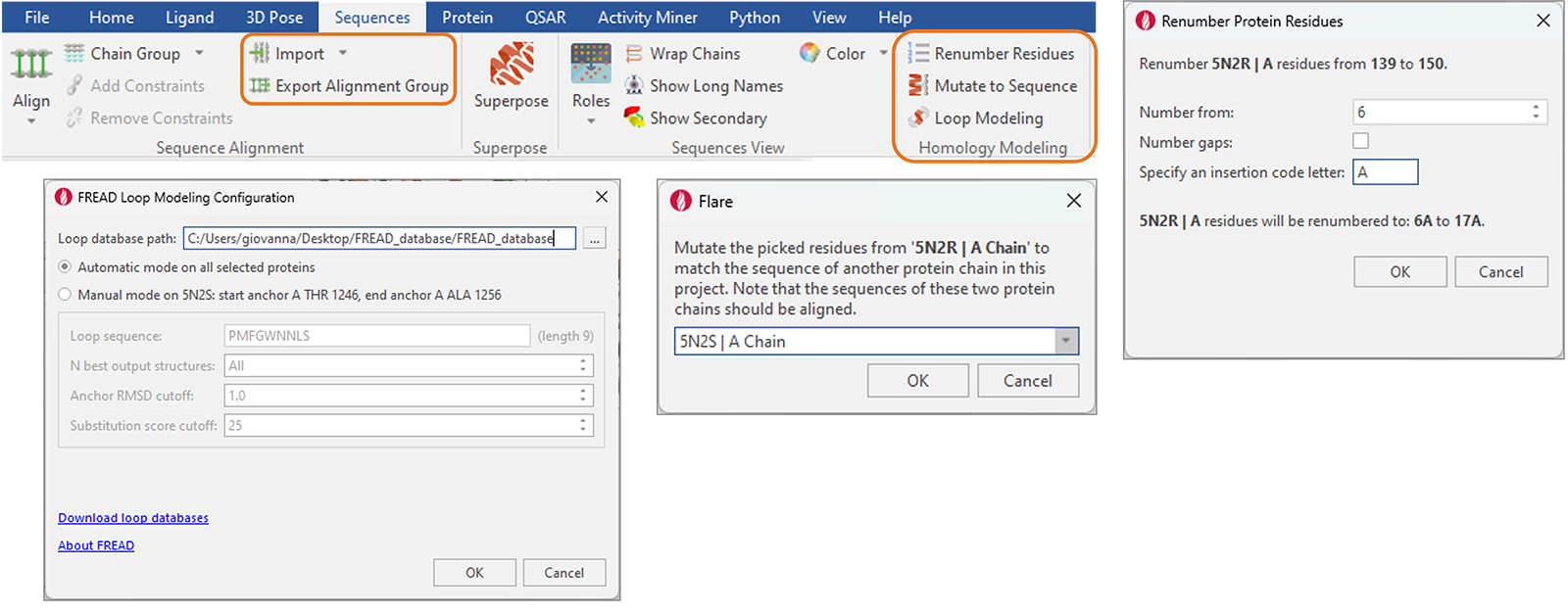
Figure 14. New and enhanced functions for protein and homology modeling are available from the reorganized ‘Sequences’ tab in Flare.
Finally, the Alignment table features a new 'Frequency Logo' to visually map consensus and diversity of sequence alignment (Figure 15): a tooltip shows all the residues which are seen in a certain position of the aligned sequences, and their frequency.
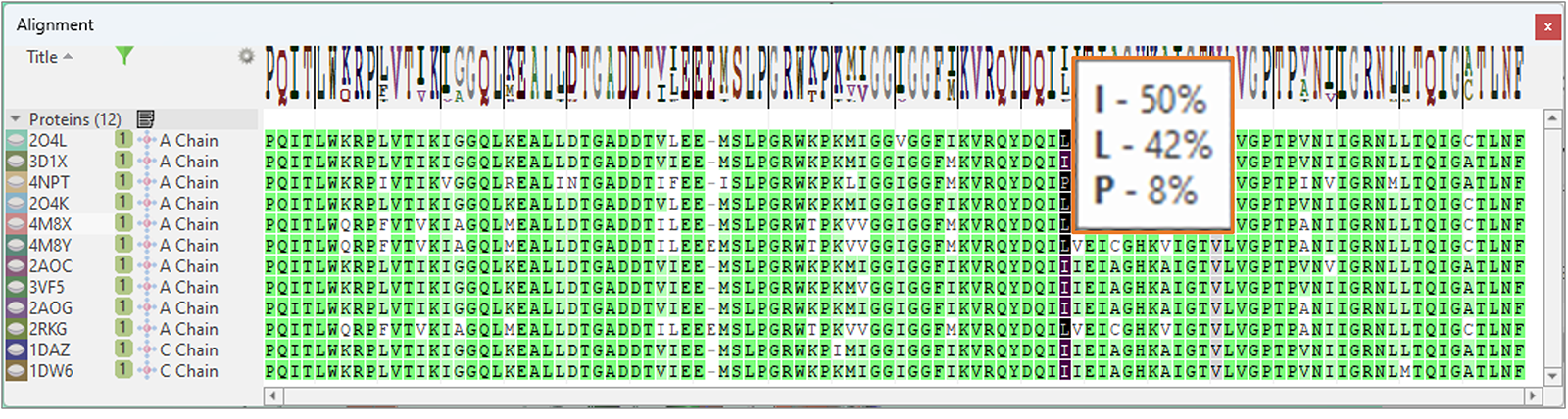
Figure 15. The new 'Frequency Logo' visually maps consensus and diversity of sequence alignment.
GIST water analysis experiments in this release can benefit from enhanced water sampling with Grand Canonical Nonequilibrium Candidate Monte Carlo (GCNCMC),6,7 especially useful to ensure optimal hydration of cryptic pockets in the protein active site.
As shown in Figure 16, GCNCMC steps can be taken during the equilibration step of the calculation, in the region defined by the GCNCMC sphere centered on the ligand included in the experiment. Size of sphere and frequency of GCNCMC moves can be fine-tuned in the advanced options.
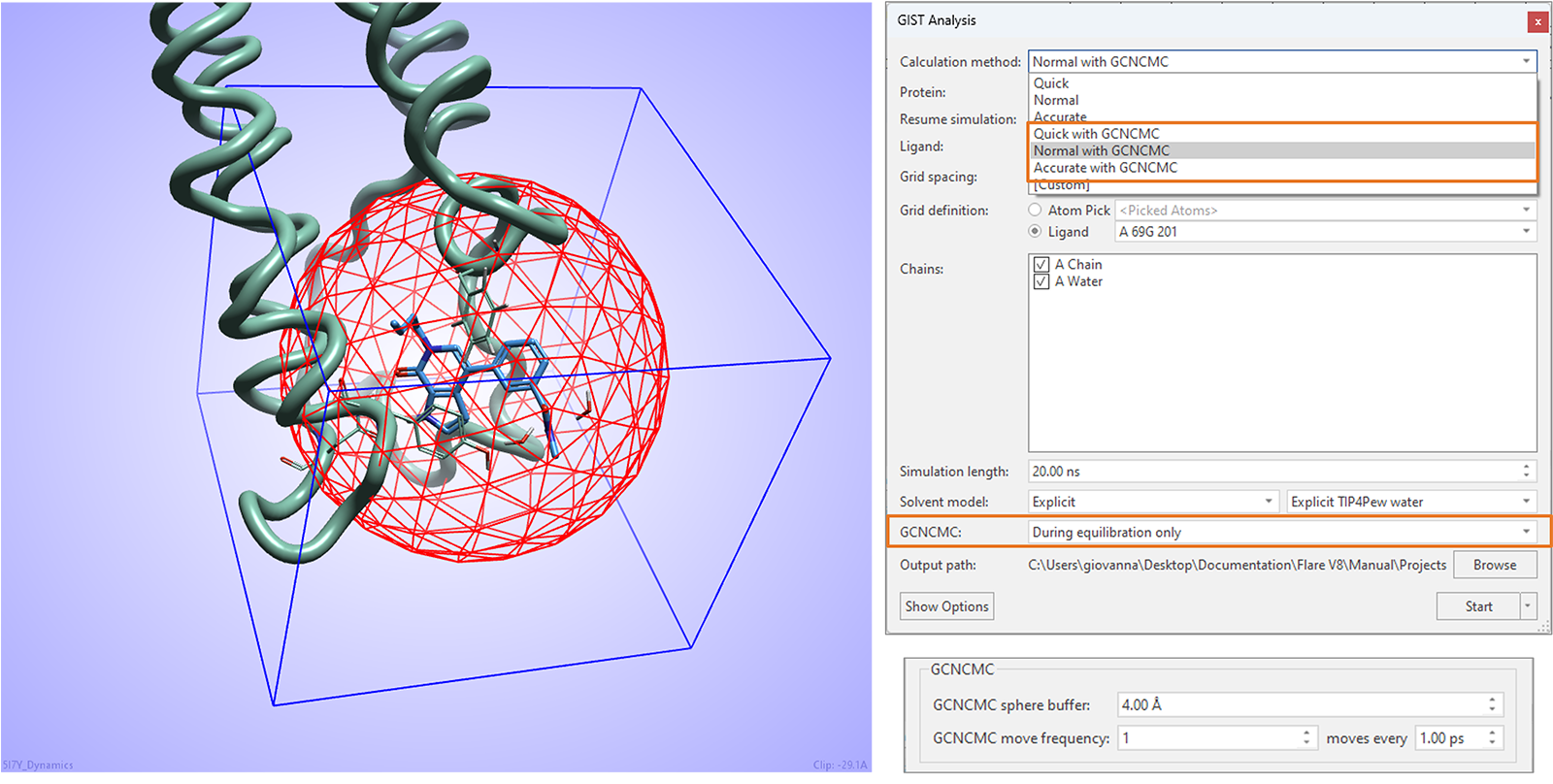
Figure 16. GIST water analysis in Flare V8 can benefit from GCNCMC moves taken during the equilibration stage of the experiment.
This new feature in Flare can be used to customize the Flare ribbon tab menus, for example by choosing one of the default profiles (Figure 17) to match your current Flare license. In this way, tabs and functions (buttons) which are not available in your current license will be hidden: furthermore, the chosen profile will be remembered as you open new projects or restart Flare. At any time, if you want to know which exciting Flare features you may be missing, choosing the ‘full’ profile will bring back all the Flare tabs and buttons.
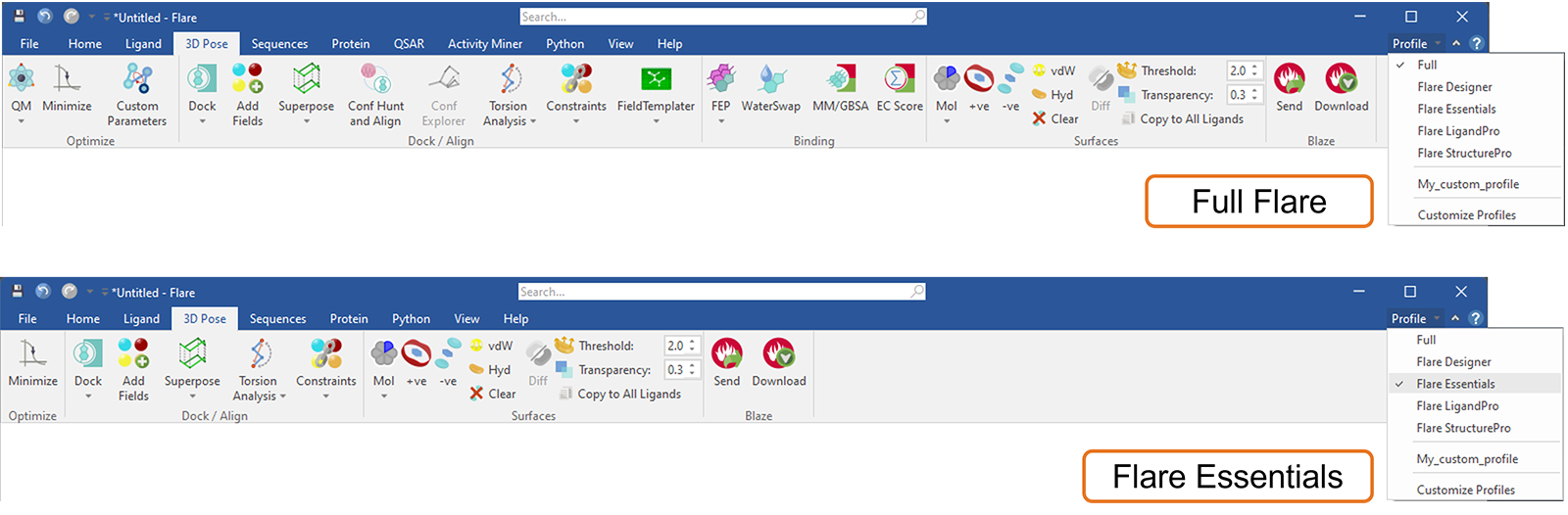
Figure 17. Choosing the Flare Essentials profile will customize the Flare menus to show only the tabs and buttons which are included in this licensing level.
You can also go one step further and create a custom profile showing only the tabs and buttons which you use more frequently, hiding all the rest, customizing their appearance (Figure 18).
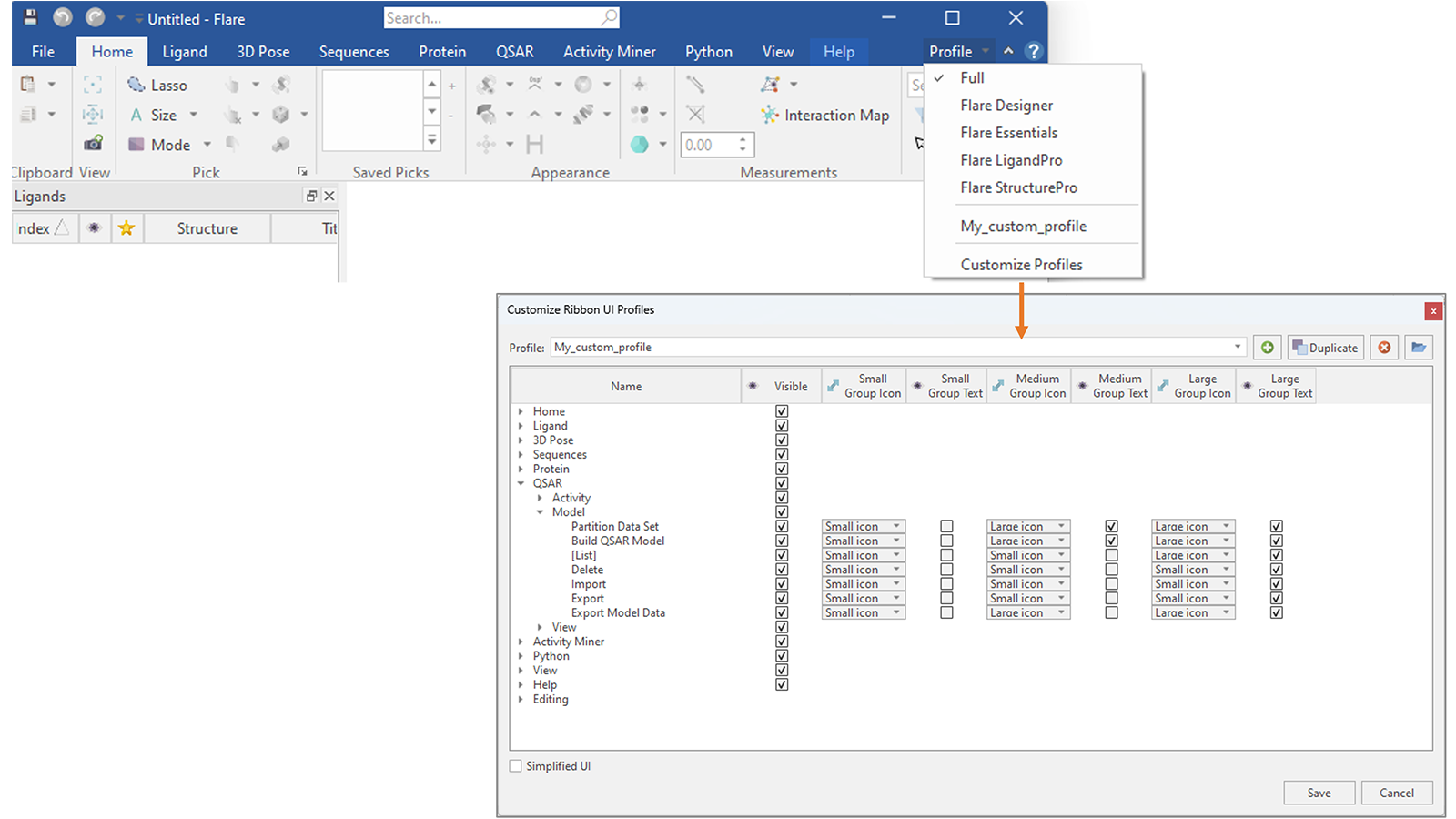
Figure 18. In Flare V8 you can create your own custom Profiles, deciding which are the tabs and buttons you want to show in the Flare menu, as well as changing their appearance.
New enhancements and improvements in Flare V8 also include:
To try out these new enhancements and the full functionality of Flare on your projects, request an evaluation and try Flare V8 today. With a range of licensing options available, Flare offers flexible solutions to meet the needs of computational chemists, medicinal chemists and academic researchers. Our dedicated support team will help you get started, with support during installation and setup, and help in accessing Flare’s wide range of features. You will benefit from full support during the evaluation process, while having the freedom to publish the results produced and use these for further research.