Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Cresset’s Spark1 software for bioisosteric replacement was used to carry out a water displacement experiment starting from the X-ray crystal structure of a selective inhibitor of Bruton’s tyrosine kinase2. The use of databases derived from available reagents ensured that the results could be tethered to molecules that were readily synthetically accessible. The availability of a sufficiently diverse source of reagents was crucial in demonstrating the feasibility of this approach.
Bruton’s tyrosine kinase (Btk) is a member of the Tec family of non-receptor tyrosine kinases. Recent literature findings2 indicate that Btk inhibition could be an attractive approach for the treatment of autoimmune diseases such as rheumatoid arthritis, a progressive autoimmune disease characterized by swelling and erosion of the joints3.
A fragment-based drug design approach was recently2 applied to the discovery of non-covalent, potent inhibitors of Btk inhibitors with Lck selectivity (Lymphocyte-specific protein tyrosine kinase, a target playing a key role in T-cell activation).
Among the most interesting hits identified with this approach, compound 2 (Table 1) was selected for further optimization. Position 8 of the cinnoline ring of fragment 2 was explored using the Suzuki−Miyaura4 synthetic methodology, starting from a series of monocyclic boronic acids/esters. This initial SAR exploration led to the discovery of compound 8 (Table 1), which shows improved potency and selectivity with respect to fragment 2.
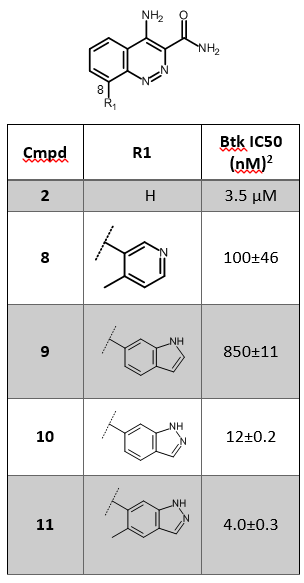
The published X-ray crystal structure of compound 8 in the active site of Btk (PDB 4ZLZ) shows a water-mediated hydrogen bond from the pyridyl nitrogen to the P-loop backbone residues Phe413 and Gly414 of Btk2 (Figure 1 – left). The replacement of 4-methylpyridin-3-yl in compound 8 with small bicyclic heterocycles displacing the water molecule and making direct H-bond interactions with the P-loop led to the discovery of compounds 10 and 11 (Table 1), with a 10-fold improved potency towards Btk.
The 3D structure of compound 8 and the bridging water molecule were used as the starting point for this Spark case study. The aim of this experiment is to verify whether our methodology is able to displace the bridging water molecule and correctly identify the same alternative indazole fragments.
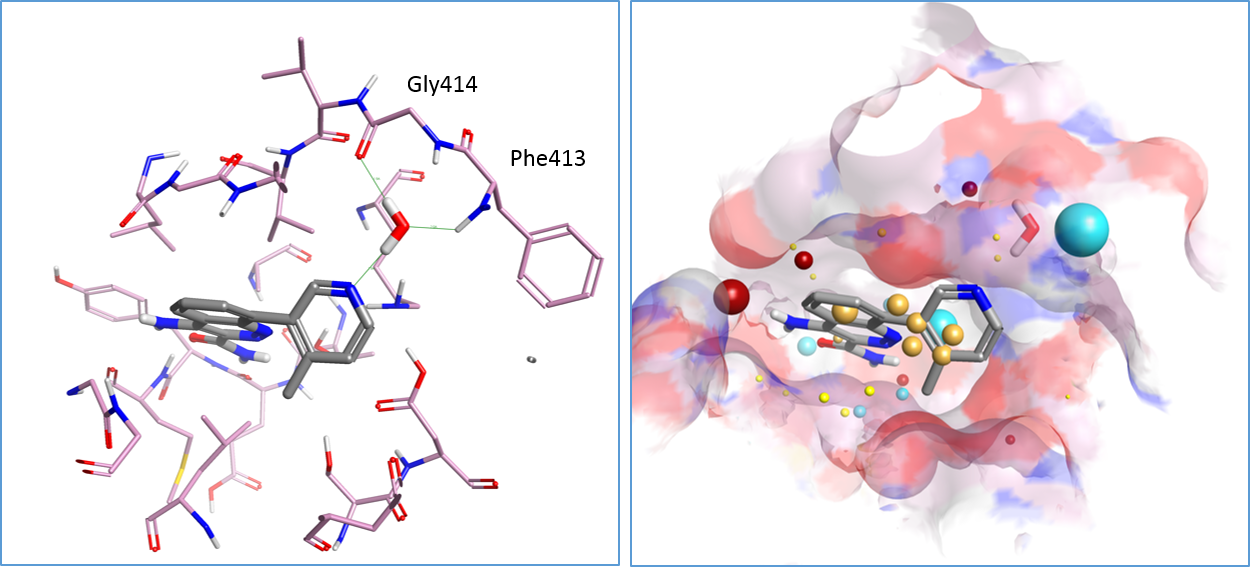
Spark’s approach to scaffold hopping and R-group replacement uses Cresset’s field-based technology5 6 to identify viable replacements for a selected portion of a reference compound using a series of fragments. In this case study we chose to use standard reagent databases7 supplied by Cresset which are based on the available chemicals directory. This gives the opportunity to rapidly search all R-groups that could be introduced at a selected position. However, an optional Database Generator module enables the creation of fragment databases that are derived from corporate compound registries or inventory systems, linking your available chemistry directly to the Spark experiment.
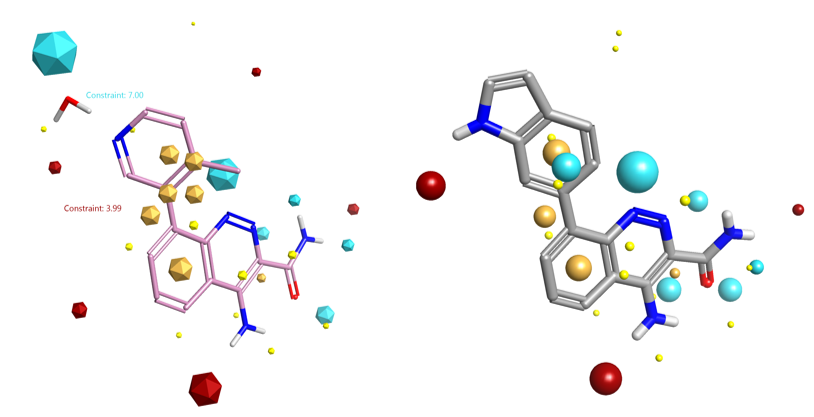
The published X-ray crystal structure of compound 8 bound into to the active site of Btk (PDB 4ZLZ) was downloaded into Forge8. The structure of the ligand was minimized and then combined with the water molecule mediating the H-bond interaction with the P-loop backbone residues of Btk to make a single molecule entry. The merging of the two 3D structures was done using the ‘combine selected pair into single molecule’ feature available in Forge. The unique entry thus created (see Figure 1 – right) was used as the Starter molecule for the Spark experiment (Figure 2 – left).
In this water displacement experiment, we want the Spark search to be driven mainly by the electrostatic fields, rather than by the usual combination of fields and shape.
For this reason a constraint was added to the negative and positive field points of the water molecule using the Spark Field Constraints Editor (Figure 2 – right). This introduced a score penalty for those results that did not match the constrained field points.
Furthermore, the ‘Normal’ conditions for scoring the Spark search results were fine-tuned to 90% Field and 10% shape, using the Btk protein as a ‘hard’ excluded volume, to constrain the size of the potential replacement fragments.
Finally, to focus the experiment on small bicyclic heterocycles, monocyclic fragments were filtered out from the list of potential results using an appropriate SMARTS filter.
Two runs of Spark were carried out using the above conditions. The initial experiment was run on a database of 775 boronic acids to closely replicate the chemistry used in the original publication2, 4.
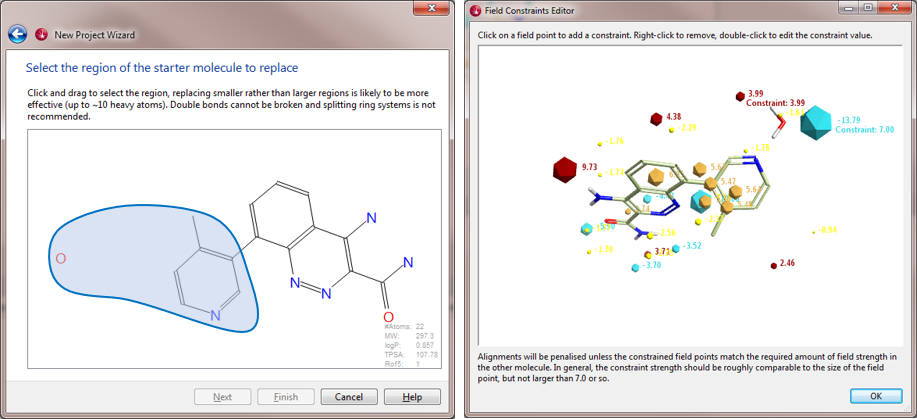
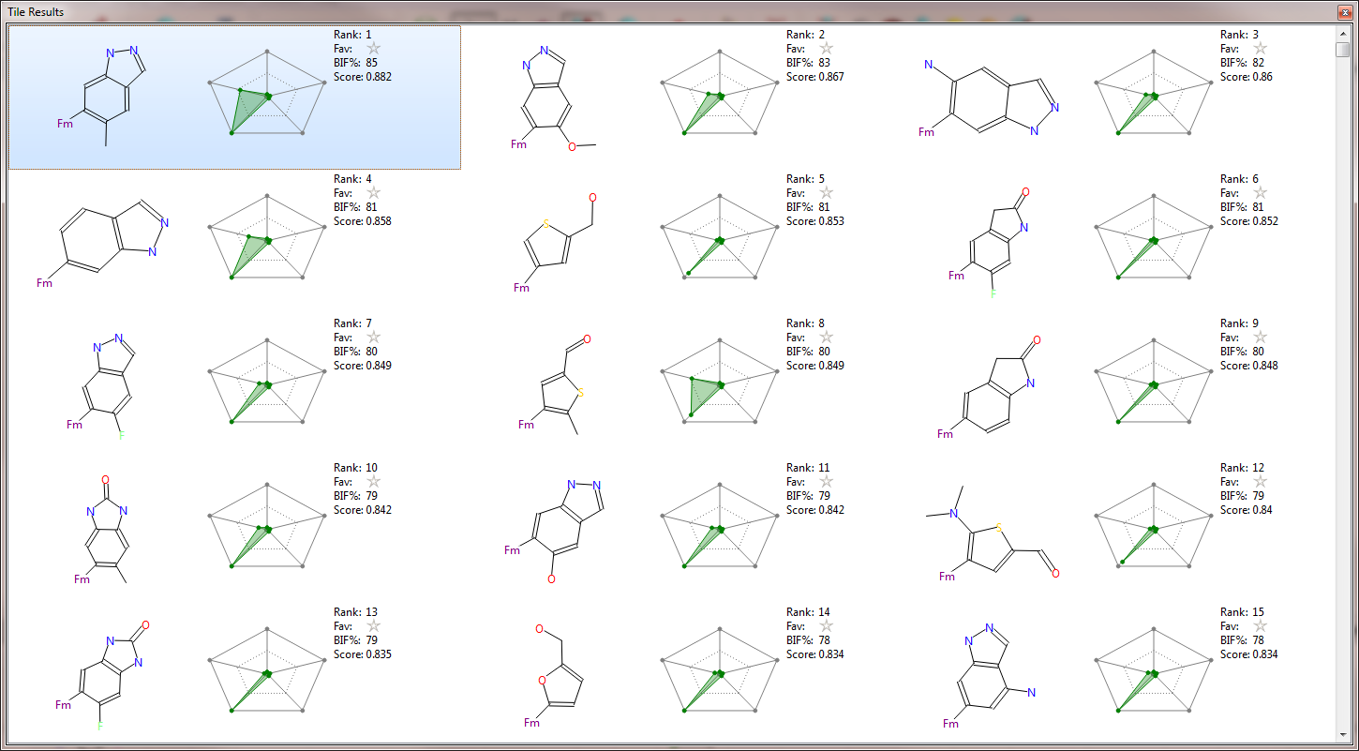
The top scoring compound from the initial search (boronic acids only) is compound 10 (Table 1). As can be seen in Fig. 3 – right, this compound superimposes very well with the starter molecule and matches the constrained field points in a satisfactory manner. However, compound 11, which would presumably superimpose even better with the conformation of the ortho-methyl-pyridin-3-yl group of compound 8, was not found in this search, due to the limited chemical diversity of the database searched.
In the second Spark search, which was run on a much larger collection of reagents (boronic acids and aryl halides), compound 11 (Fig. 3 – center and Fig. 4) is the top scoring result, while compound 10 ranks 4th in the list (Fig. 4).
The original paper2 also reports the indole-substituted compound 9 (Table 1), quite similar in terms of 2D structure to the much more potent indazole compounds 10 and 11. This fragment is available in both the databases searched, but is not retrieved by Spark. The indole fragment in fact cannot match the constrained negative field point of the bridging water molecule, as shown in Fig. 5, where compound 9 is shown superimposed to the starter molecule in Forge. The lack of this relevant interaction explains the much lower potency of compound 9, with a Btk IC50 = 850nM (Table 1).
Figure 4 shows a tile view of the 16 top scoring results from the second Spark experiment. Several different flavors of the indazole fragment carrying different substitution patterns are represented in this list. Alternative bicyclic fragments are also proposed, which may provide useful ideas for a further exploration of this target.
In this case study Spark successfully managed to displace the crystallographic water molecule bridging the interaction between compound 8 and the P-loop of Btk, replacing it with small, synthetically accessible bicyclic heterocycles.
Availability of appropriate sources of chemical diversity is still a key factor in determining the success of any bioisosteric replacement experiment.
For this reason, the creation of fragment databases derived from corporate compound registries or inventory systems, linking your available chemistry directly to the Spark experiment, is highly recommended.
1. http://www.cresset-group.com/products/spark/
2. Smith, C. R. et al., J. Med. Chem. 2015, 58, 5437−5444
3. Firestein, G. S., Nature 2003, 423 (6937), 356−361
4. Miyaura, N., Suzuki, A. et. al., J. Am. Chem. Soc. 1989, 111 (1), 314−321.
5. J. Chem. Inf. Model., 2006, 46, 665-676.
6. http://www.cresset-group.com/science/field-technology/
7. Spark fragment databases come from commercial compounds, ChEMBL, ZINC and VEHICLe.
8. http://www.cresset-group.com/products/forge/