Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
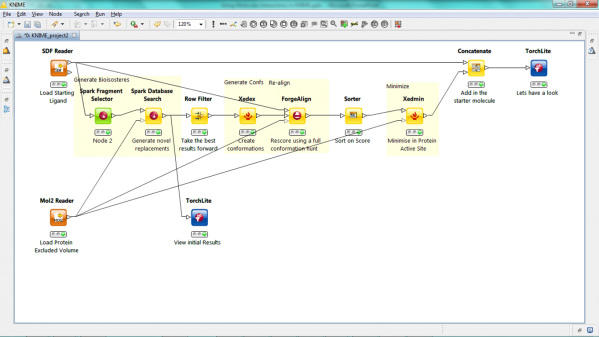
We have released a new version of our KNIME nodes that dramatically increase our offerings on this growing platform. We have added Spark and the 3D-QSAR functionalities of Forge resulting in 6 new nodes, 10 in all. Lastly we have expanded the capabilities of the XedMin node to enable minimization of ligands within a protein cavity and of our viewer node to accept TorchLite, Torch or Forge binaries as the viewing application.
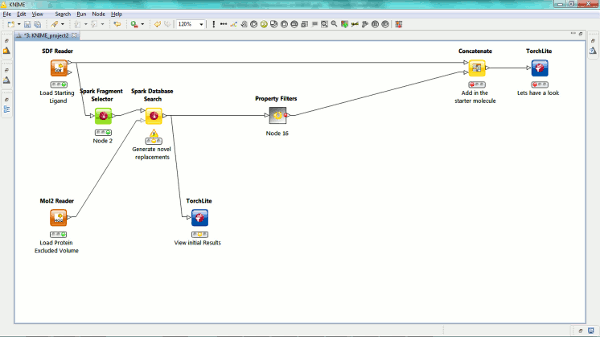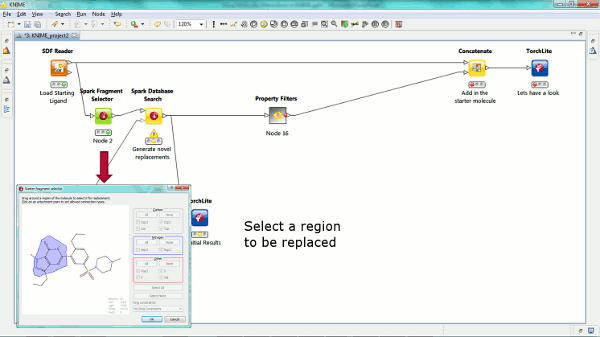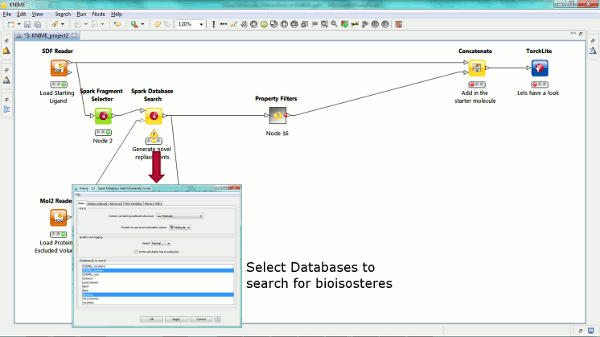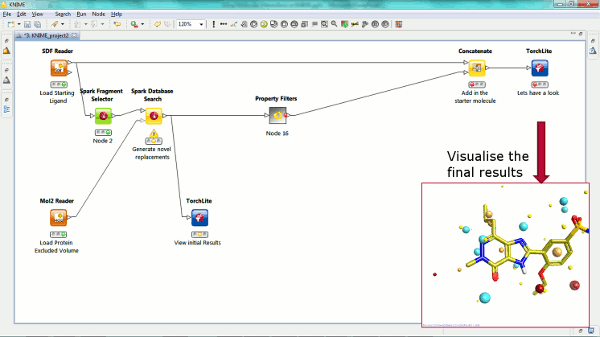
Our new Spark nodes enable the rapid generation of new bioisosteric scaffolds from each molecule that is input. Using Spark in a KNIME workflow enables the assessment and clustering of the results using any of the myriad of methods that are available in this environment. Equally the results could be further analyzed using other Cresset nodes such as the new ability to minimize inside the protein cavity using XedMin.
The ligand alignment method of Forge has been available in KNIME for some time. In this release we build on this capability to add all the remaining Forge functionality. In particular the new ‘Forge_build’ node enables the building of 3D-QSAR model using our Field-point sampling method and the scoring of new molecules against these models using the new ‘Forge_score’ node. The Forge_build node uses our in-built PLS algorithm to find the relationship between field values and activity but in the KNIME environment the raw data can be output and analyzed using other KNIME methods such as random forests.
In this release we have updated the XedMin node to enable minimization of a ligand in a protein active site. The protein file is loaded as an extra input and held static in the minimization enabling each ligand in the KNIME table to be individually minimized. Adding this to the result of a Spark or Forge alignment node gives an optimized view of new scaffolds or ligands.
We have provided a molecule viewer node in KNIME for a couple of years. Now we have updated the node to enable the node to call TorchLite, Torch, or Forge applications to view the results of any molecular manipulation or Forge project file.
Lastly we have changed how we deliver our KNIME nodes. To get the nodes simply point KNIME to the following installation site: http://www.cresset-group.co.uk/knime/update. Detailed instructions for this are contained in the latest release notes. Remember the nodes themselves are not functional without access to the appropriate command line binary. You can get an evaluation of the command lines from http://www.cresset-group.co.uk/cgi-bin/demo.cgi.