A three way collaboration between Cresset, the University of Newcastle and Sygnature Discovery
Research and development is not an easy business. Real research invariably involves starts and stops, misassumptions, poor data and ill-defined targets. The resulting path is often convoluted, peppered with roadblocks and pitfalls and at best is a fairly untidy process.
This is an account of a real project, warts and all and is not untypical of projects conducted either in academia or the pharma industry. Ultimately, progress was made and knowledge was gained as new data from the biology, chemistry and modeling were carefully applied on the way.
In this case, despite the difficulties described below, the project enjoyed a relatively rapid progression from a patent bust to a novel and selective series with many of the problems associated with the target ultimately solved. The project is on track and Cresset’s modeling has provided unique and valuable insights and will continue to add value as it is applied moving forwards.
The project
From 2012 to 2014 Cresset consulting services contributed computational modeling effort to a three-way collaboration with the University of Newcastle and Sygnature Discovery on an MRC funded osteoarthritis project. Strathclyde1 and Newcastle2 Universities conducted vital experiments that identified the protein matriptase as a key mediator in disease related pathways leading to collagen degradation in joints.
This protein target3, a member of the Serine protease family, had been crystalized with peptidomimetic inhibitors adapted from a urokinase inhibitor series by Steinmetzer4. These contained the well-known benzamidine P1 pocket interacting groups which are critical for potency in many of this class of protease.
The aim of the modeling exercise was to provide Newcastle University with a new lead series of inhibitors of matriptase, with both an IP position and importantly a more favourable profile for delivering in-vivo activity.
Method
Read the details in the full write-up.
In brief, the XED force field was used to model the binding hypothesis for a potent ligand. This large reference compound was split into three appropriately sized, and more lead-like molecules as shown below, and used as the input for a Blaze virtual screen in order to find alternative scaffolds.
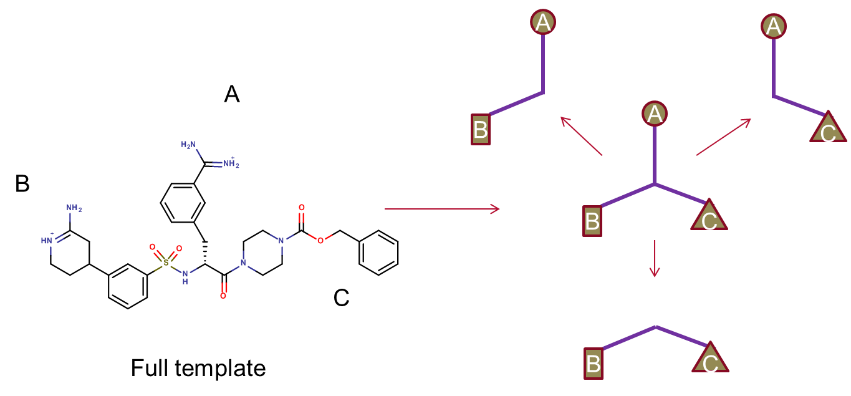
Figure 1. Fragmentation strategy for Blaze search molecules.
Unfortunately, only a small proportion of the virtual screening output was ultimately purchased and screened and of a 142 molecules only a single example was found (Compound 0154) which had appreciable activity at matriptase – albeit relatively weak (IC50=30 μM).
A parallel patent busting strategy
At this time the chemistry partner had put in place parallel efforts on a patent busting strategy from a recently reported synthetic inhibitor. The disadvantage of this approach was that the patent bust was from a symmetric tribasic molecule and so optimisation, particularly of the ADMET profile, would be problematic. Furthermore, the binding hypothesis modeling was made more complex by the inherent ambiguity provided by the symmetrical starting ligands.
Read more in the full write-up.
This parallel strategy provided some comparable actives to Compound 0154 but via a far more expedient chemistry and with potential to design-in an IP position.
Final state of play
The project enjoyed a relatively rapid progression from a patent bust to a novel and selective series with many of the problems associated with the target ultimately solved. One exception was the main liability issue of ‘membrane penetration’ which was present from the outset and remains to be resolved. But, with one potent series well established, and a second series from the virtual screen with potential to be explored, the project is on track to provide what was promised.
Cresset’s modeling provided unique and valuable insights to the project and will continue to add value as it is applied moving forwards towards the development of series 1 and 2.
Acknowledgements
Sygnature (chemistry): L. Duffy, P. Meghani.
University of Newcastle (biology): Prof. Drew Rowan, W. Hui, D. J. Wilkinson, A. Destrument, S. Watson.
Cresset (modeling): A. Vinter.
References,
- Ferral, W. R. et al, Protease-activated receptor-2: a novel pathogenic pathway in a murine model of osteoarthritis, J. Arthritis & Rheumatism, 2010 (http://strathprints.strath.ac.uk/20137/).
- Rowan, A. D. et al, Matriptase Is a Novel Initiator of Cartilage Matrix Degradation in Osteoarthritis, Arthritis & Rheumatism, 2010, Vol. 62, No. 7, pp 1955–1966.
- http://merops.sanger.ac.uk/; search: ID = S01.302: Name = matriptase.
- Steinmetzer, T. et al, Secondary Amides of Sulfonylated 3-Amidinophenylalanine. New Potent and Selective Inhibitors of Matriptase, J. Med. Chem. 2006, 49, 4116-4126.
