There has been a revival of interest in covalent inhibitors in recent years. This has culminated in a growing list of diverse proteins that have been successfully targeted, that are outside the well-trodden protease area.1-4
An older non-protease example is Clopidogrel. This P2Y12 inhibitor requires activation in vivo (through oxidative metabolism) to generate the electrophile intermediate (sulfenic acid) that forms a covalent disulphide bridge to a cysteine residue in the target GPCR. Unravelling the true mechanism of this compound led to second generation inhibitors which further exploit the covalent mode of action5.
Potential benefits
One more recent, and well documented, non-protease example is in the field of oncology, with the drug Ibrutinib. The covalent binding of the inhibitor to the target kinase BTK demonstrates multiple benefits:
- It removes competition with the endogenous ligand, as is true for other covalent inhibitors, but in this case is more significant as concentration of competing cellular ATP is in the millimolar range.
- High efficacy, in contrast to some conventional inhibitors, gained specifically by using this covalent inhibitor design.
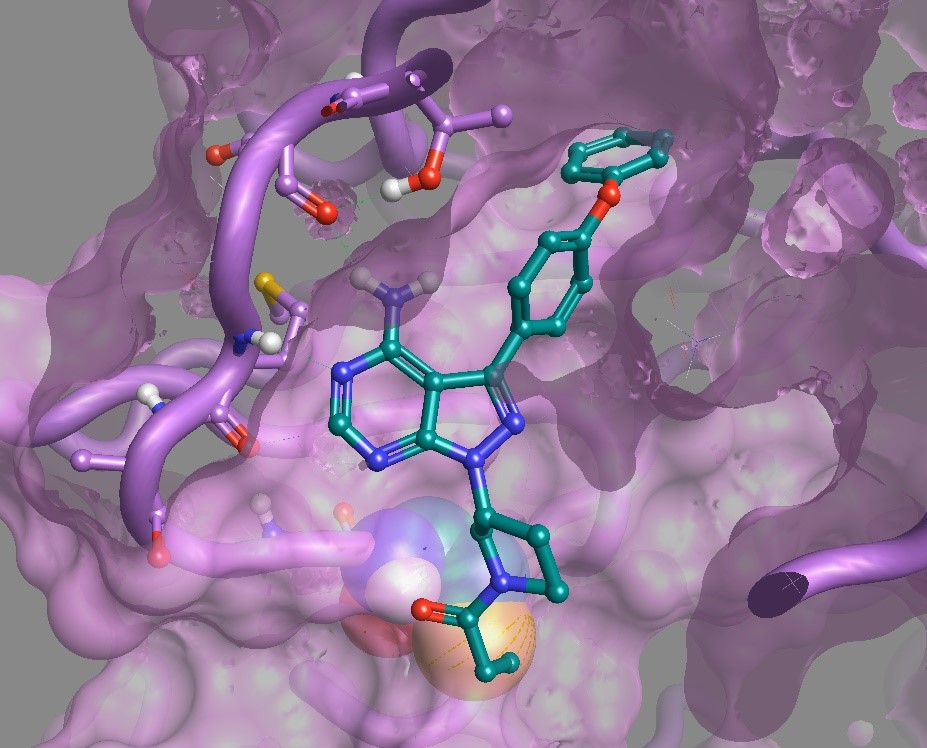
Figure 1: Ibrutinb in BTK PDB: 5P9J showing Cys-481 spheres near with the covalent linkage visualized using Flare a new structure-based design application from Cresset.6
From a safety point of view, a method of achieving high efficiency/potency of any drug should translate into a lower dose that is ideally below any potential toxicity threshold (very few drugs are given at =< 10mg per day!). Similarly, prolonged duration of action, a consequence of the covalent inhibition, can result in less frequent dosage regimes that could be very beneficial – depending on the exact pharmacology.
So why have drug discovery companies not adopted this approach more broadly?
Mainly, because this is not a panacea – some proteins are more quickly processed, turned over or are mutated. The BTK case itself is an unfortunate example of the latter; the resistance mutation C481S produces a protein that cannot undergo the reaction with the inhibitor and ultimately limits its utility in patients. What may have more generally hampered covalent inhibition strategies in the past, observed particularly in the field of protease inhibitor design, was the high electrophilicity and poor selectivity. The very justifiable fear of off-target activity and threat of unresolvable toxicity issues, has contributed to the adoption of a dogma which still significantly disfavors the approach across the industry. The lack of a wider uptake of the approach since then has been exacerbated, perhaps, by the industrialization of experimental design and/or the lack of attention to the kinetic behavior of these systems. One size doesn’t really fit all!
Computational chemistry is critical for modern covalent inhibitor design strategies
An appreciation of the kinetics of molecular recognition and its impact on drug efficacy (Figure 2) has had a resurgence; together with the introduction of biophysical techniques, such as Surface Plasmon Resonance (SPR), which allow the routine resolution of otherwise painfully elusive kinetic parameters such as kon/koff.
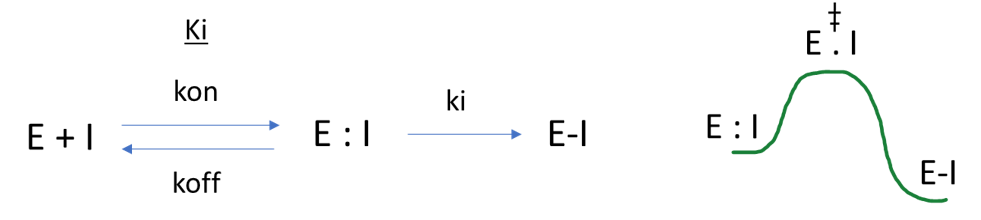
Figure 2: Schematic of the covalent inhibition process (ki/Ki equivalent to classical Michaelis Menten enzymic parameters kcat/Km), transition state energy diagram for the covalent transformation.
The observed rate of inhibition for a covalent inhibitor is effectively dependent on the ratio of the rate constant for the chemical transformation of the inhibitor (ki), relative to the rate of unbinding (koff). We can introduce a level of catalysis in this system if we design appropriate geometries that lower the energy of (i.e. favor) the E : I complex7. If we are talking about hand-crafting optimum molecular geometries, then we are usually thinking about utilising computational chemistry as a technique.
Advances in the covalent inhibition area have clearly arisen from a systematic investigation into the modulation of the intrinsic reactivity of these electrophilic ligands. Covalent inhibitor design, in its more current form, now reduces reliance on the Ki, and drives optimization through binding (i.e., reducing koff) rather than reactivity. This more thoughtful approach is an essential step towards maximizing selectivity and minimizing off target activity. In parallel with the ‘much needed’ experimental investigations, a range of computational techniques are required for modern covalent inhibitor design strategies. These include:
Ligand-based design
- Virtual calculations defining reactivity and ranking of electrophiles.
Structure-based design
- Optimizing the molecular recognition toward the target enzyme by careful design of the ligand functionality through steric and electrostatic control.
- Optimizing placement of an appropriate electrophile on the inhibitor in relation to an accessible, non-conserved nucleophile on the protein. Selectivity of the covalent bonding step is governed by the choice of a suitable electrophile and proximity to the nucleophile. Optimizing the reactivity of the electrophile for a covalent inhibitor requires a balance between achieving a sufficient high rate of reaction with the selected nucleophile on the target protein as well as minimal rate of reaction with off target nucleophiles.
We anticipate that this approach will prove increasingly useful, particularly for otherwise intractable biological targets involving large and complex protein-protein contacts.
Add direction and insight to your projects
Cresset Discovery Services’ experienced scientists work alongside your chemists to solve problems, provide fresh ideas, remove roadblocks and add direction and insight to your projects.8 They offer expert molecular design capabilities and use cutting-edge technologies to provide support for numerous client projects, across a range of disciplines.
Free confidential discussion
To find out how Cresset Discovery Services can help you reach your next milestone faster and more cost effectively, contact us for a free confidential discussion.
References and links
- Bauer R.A., Covalent inhibitors in drug discovery: from accidental discoveries to avoided liabilities and designed therapies, Drug Discov. Today, 2015, 20 (9), 1061-1073.
- Baillies T.A., Targeted Covalent Inhibitors for Drug Design, Angew. Chem. Int. Ed., 2016, 55, 13408-13421.
- De Cesco S., Kurian J. Dufresne C., Mittermaier A.K., Moitessier N., Covalent inhibitors design and discovery, Eur. J. Med. Chem., 2017, 138, 96-114.
- Lagoutte R., Patouret R., Winssinger N., Covalent inhibitors: an opportunity for rational target selectivity, Curr. Opin. Chem. Biol., 2017, 39, 54-63.
- Exploring synthetically accessible alternatives to P2Y12 antagonists using electrostatics and shape.
- https://www.cresset-group.com/flare/
- See ‘Hammond postulate‘ for an exothermic reaction.
- https://www.cresset-group.com/discovery-services/

