Events
Symposium on Streamlining Drug Discovery, San Diego
Find out about new approaches and technologies being applied to the search for future therapeutics.
This free to attend, one day symposium, is aimed at scientists from pharmaceutical, biotechnology, agrochemical, flavor and fragrance organizations, not-for-profits and academia who wish to get a broad appreciation of the latest advances in drug discovery delivered by key scientists and thought leaders from leading organizations.
Other dates and locations
If you are unable to attend this symposium, you or your colleagues may be interested in the events taking place on:
October 18, Cambridge MA
October 25, San Francisco CA
| 09:30 | Registration | |
| 10:00 | Welcome from meeting chairman | Hanneke Jansen, Novartis |
| 10:05 | Keynote: The Analysis-Knowledge-Action Principle in Drug Discovery: A Focus on Quality | Hanneke Jansen, Novartis |
| 10:50 | In Silico and In Vitro Assessment of OATP1B1 Inhibition in Drug Discovery | Matt Danielson, Lilly |
| 11:20 | Break | |
| Using Water Swap to Assess Target Druggability | Adam Kallel, Retrophin | |
| 12:20 | Electrostatic Complementarity™ as a New Approach to Visualize and Predict Activity | Sylvie Sciammetta, Cresset |
| Lunch | ||
| Advancing Therapeutics for Opportunities in Medicine | Jim Brase, Lawrence Livermore National Laboratory | |
| 14:15 | Imputation of Protein Activity Data Using Deep Learning | Matthew Segall, Optibrium and Tom Whitehead, Intellegens |
| Break | ||
| Medicinal Chemist’s Relationship with Additivity: Are we Taking the Basics for Granted? | Guy Breitenbucher, UCSF | |
| 15:45 | Managing External Chemistry for Effective Support of Drug Projects | David Hollinshead, Elixir Software and Andrew Griffin, Praxis Precision Medicines |
| 16:15 | Closing remarks | Hanneke Jansen, Novartis |
| Drinks reception |
The Analysis-Knowledge-Action Principle in Drug Discovery: A Focus on Quality
Hanneke Jansen, Novartis
Computer-aided drug discovery is most successful when designs and hypotheses fully leverage the existing knowledgebase while at the same time creating opportunities for serendipitous discoveries and exploration of new chemical space. When venturing into the unknown, the knowledge we seek to apply is aimed at improving the quality of our designs and hypotheses. Improving quality means different things for different projects and for different stages of the same project. After defining the quality dimension that is relevant for a given project, we apply the Analysis-Knowledge-Action, or AKA, principle: We analyze the relevant data, extract knowledge, and formulate an explicit action. This lecture will describe application of our AKA principle in designing compound sets for screening and in generating medicinal chemistry design principles for optimization.
The quality dimension that will be highlighted in the compound set design application relates to cytotoxicity. Analysis of our in-house data allowed us to extract knowledge, initially in the form of a logD-driven relationship to cytoxicity, for application in antibacterial projects. Subsequent work resulted in a predictive model using the pQSAR platform that leverages the entire Novartis knowledgebase and provides cytotoxicity risk assessment for drug discovery in all disease areas. This model can be used in the explicit action of biasing compound set designs when selecting compounds from compound archives, hit-lists, or virtual libraries. A customizable KNIME workflow will be described that allows this and other quality measures to be balanced with optimal coverage of diversity for compound set designs.
The optimization application will show the use of matched molecular pair (MMP) analyses to extract knowledge in the form of design principles related to quality measures for in vitro ADME endpoints. The MMP analysis across our whole corporate archive allows identification of transformations that consistently improve the quality of the compound in the selected dimension. The explicit action in these examples is the synthesis of selected compounds. Computational tools that are needed include analyses platforms and visualizations that function as idea generators and allow team members to interpret the likelihood that certain transformations have the desired impact, thus inspiring compound synthesis.
In Silico and In Vitro Assessment of OATP1B1 Inhibition in Drug Discovery
Matt Danielson, Lilly
The organic anion-transporting polypeptide 1B1 transporter belongs to the solute carrier superfamily and is highly expressed at the basolateral membrane of hepatocytes. Several clinical studies showed drug-drug interactions involving OATP1B1 thereby prompting the International Transporter Consortium to label OATP1B1 as a critical transporter that can influence a compound’s disposition. To examine OATP1B1 inhibition early in the drug discovery process, we established a medium-throughput concentration-dependent OATP1B1 assay. In order to create an in silico OATP1B1 inhibition model, deliberate in vitro assay enrichment was performed with publically known OATP1B1 inhibitors, non-inhibitors, and compounds from our own internal chemistry.
In this presentation the strategy and results of assay enrichment, along with the performance of subsequent QSAR model(s) will be discussed. In addition to the in silico OATP1B1 inhibition QSAR models, physicochemical trends were also examined to provide structure activity relationship guidance to early discovery teams.
Managing External Chemistry for Effective Support of Drug Projects
David Hollinshead, Elixir Software and Andrew Griffin, Praxis Precision Medicines
The pharmaceutical and biotechnology R&D model for in-house drug projects, and their chemistry components, has evolved over recent decades. The drivers for conducting projects through collaborations using dispersed global teams are readily apparent. Yet various challenges in managing the essential, iterative design – make – test – analyze cycle, have persisted.
This joint presentation highlights the nature of the challenges faced, and provides a case history of how iterative design – make – test – analyze cycles can be efficiently delivered through an intuitive, real-time software platform which enables a co-ordinated chemistry contribution to drug projects.
Using Water Swap to Assess Target Druggability
Adam Kallel, Retrophin
Only 2% of human proteins interact with approved drugs. It is also estimated that only about 15% of human proteins are disease modifying and only about 12% are druggable (with no correlation between the two sets). Consequently, only about 2% of disease modifying proteins would be druggable. I used Water Swap to assess a number of proteins that fell in to the category of having ligands that advanced into clinically evaluated drugs and those that had been studied and were not able to advance preclinical ligands into clinical evaluation. Proteins that had clinically advanced ligands were found to have pockets that had a favorable DG for ligand binding as opposed to water. Those Proteins that had not yielded clinically advanced ligands had pockets that favored water rather that ligand. Water Swap is a good method to evaluate protein druggability.
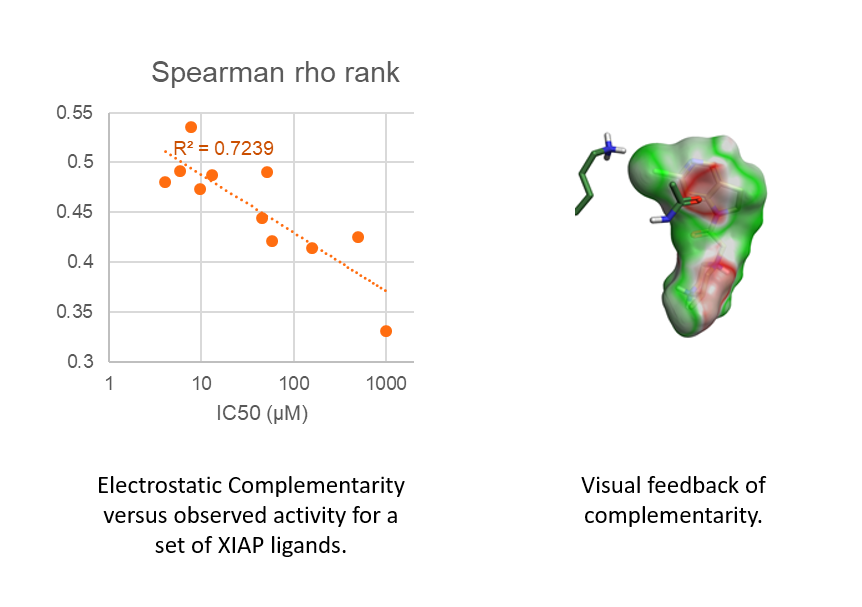
Electrostatic Complementarity™ as a New Approach to Visualize and Predict Activity
Sylvie Sciammetta, Cresset
Electrostatic interactions between small molecules and their respective receptors are a key contributor to the free energy of binding. Assessing the electrostatic match between ligands and binding pockets provides therefore important insights into ligand binding and molecule design.
The polarizable XED force field is an excellent base for calculating electrostatic properties due to its description of anisotropic atomic charge distributions and relatively modest computational costs. By computing electrostatic potentials for both ligand and protein with XED, the Electrostatic Complementarity™ (EC) of complexes can be calculated and translated into a simple coloring scheme on the ligand and protein surface.
We present the theoretical background of our EC calculations and discuss their application to mGluR5, XIAP, and other selected targets. We show how using EC can inform SAR interpretation and new molecule design.
Capturing and Applying Knowledge to Guide Compound Optimization
Matt Segall, Optibrium
Compound design requires a combination of knowledge and expertise from different perspectives: Understanding of structure-activity relationships (SAR), based on data from previously studied compounds; expertise from diverse fields to define the multi-parameter optimization (MPO) objectives of a project; and knowledge of synthetic strategies that may be applicable to create the next rounds of compounds for investigation. All of these forms of knowledge can be captured and applied computationally: Machine learning methods can generate quantitative structure-activity relationship (QSAR) models to predict the properties of novel, virtual compounds[1]; MPO methods capture the desired property criteria for a successful compound for a specific project and rigorously prioritize ideas for consideration[2]; and, optimization strategies can be captured as structural transformations that reflect steps made in previous chemistry projects[3,4].
In this presentation, we will describe these methods and illustrate how they can be seamlessly combined to rigorously explore new, relevant compound ideas and prioritize those most likely to achieve a project objective. This approach can help to stimulate the search for new optimization strategies and explore a much broader range of compounds than could be achieved based on a single chemist’s or even a project team’s experience. Example applications include the optimization of compounds with a desired polypharmacology or selectivity profile and exploration of lead hopping strategies to overcome pharmacokinetic issues, while maintaining target potency.
[1] O. Obrezanova, M.D. Segall, J. Chem. Inf. Model. (2010) 50 (6), pp. 1053-1061
[2] M.D. Segall, Curr. Pharm. Des. (2012) 18(9) pp. 1292-1310
[3] M.D. Segall et al., J. Chem. Inf. Model. (2011) 51(11) pp. 2967-2976
[4] I. Ujváry, J. Hayward, In N. Brown ed., Bioisosteres in Medicinal Chemistry (2012)
Imputation of Protein Activity Data Using Deep Learning
Tom Whitehead, Intellegens
The knowledge of compound bioactivity data against drug targets underpins the discovery of new drugs. However, databases are currently sparse; for example, the ChEMBL dataset is just 0.05% compete and the sparsity of data in proprietary pharma databases is similar. We will describe a novel deep learning algorithm to capture correlations within protein activity data, as well as between molecular descriptors and protein activities, to impute the missing activities. Unlike many deep learning methods, this approach is capable of being trained using sparse and variable data, typical of those available in drug discovery. We will present examples illustrating the application of these deep learning networks to impute missing activities in the sparse input data, as well as to make predictions for new compounds based on molecular descriptors alone. The results will be compared with conventional machine learning methods such as random forests and Gaussian processes.
Medicinal Chemist’s Relationship with Additivity: Are we Taking the Basics for Granted?
Guy Breitenbucher, UCSF
Given the importance and complexity of ligand-protein interactions in drug discovery, medicinal chemists continuously rely on structure activity relationships to design molecules which improve binding affinity. Crucial to predicting activity is an understanding of additive relationships in the SAR. Yet most drug programs never quantitate the additive SAR relationships when working on drug discovery programs, even in an environment that has made the synthesis of matrix libraries relatively simple. Presented here will be simple to use methods for quantitating additive relationships within SAR matrices and a number of examples of the utility of these methods in drug discovery projects. In addition, an SAR example which clearly confirms protein structural changes as the source of some non-additivity will be presented.
Guy Breitenbucher
Dr. Breitenbucher has over 20 years of drug discovery experience and is currently Professor at the Institute for Neurodegenerative Diseases (IND) at UCSF. Guy manages a team of drug discovery chemists within the IND focused on discovering treatments for disorders in which protein miss-folding is the primary pathological component.
Prior to joining UCSF, Dr. Breitenbucher was senior director of Chemistry at Dart NeuroSciences (DNS). Dart’s focus was discovering drugs that enhance synaptic plasticity for the treatment of a variety of CNS disorders. During his 8 years at DNS he built a dynamic scientific department that discovered 10 novel pre-clinical candidates utilizing 7 different biological mechanisms, currently 4 of these molecules are in clinical evaluation for the treatment of Parkinson’s, Stroke, and Schizophrenia.
Prior to Dart, Guy was a research fellow at Johnson & Johnson and headed Pain Discovery Chemistry. At J&J, Dr. Breitenbucher worked on or led drug discovery projects in 10 different biological mechanisms which advanced compounds into pre-clinical or clinical development for a variety of disorders. Dr. Breitenbucher received his B.Sc. and M.Sc. in chemistry at California State University at Long Beach and his Ph.D. in organic chemistry at University of California at Riverside. Dr. Breitenbucher is co-author on over 50 peer reviewed scientific papers, an inventor on over 40 patents.
Matt Danielson
Matt Danielson earned his PhD in Medicinal Chemistry and Molecular Pharmacology from Dr. Markus Lill’s group at Purdue University where he studied molecular dynamics and docking in CYP enzymes. After completing his PhD, Matt joined Dr. Michel Sanner and the AutoDock group at the Scripps Research Institute for his postdoc and worked on incorporating protein flexibility into molecular docking. In 2013 Matt joined the computational ADME group at Eli Lilly where he has supported drug discovery efforts in several therapeutic areas.
Andrew Griffin
Following a Ph.D with Tim Gallagher and postdoctoral studies with Professor Stephen Hanessian, Andrew moved to the AstraZeneca (Montreal) where he held the position of Associate Director in Medicinal Chemistry. He led multiple chemistry teams within AZ and with external chemistry partners, delivering lead generation projects and clinical candidates. Throughout his career Andrew has maintained a keen interest in using informatics to aid drug discovery and was member of the AZ Predictive Chemistry Network as well as leading a global initiative that provided a platform to facilitate external chemistry and testing. Since 2012 he has become a medicinal chemistry consultant and has been working with Praxis Precision Medicines since its creation.
David Hollinshead
David is Technical Director at Elixir Software Ltd with a primary focus customizing software applications matched to customer needs. Before Elixir was founded in 2012, David spent 28 years within AstraZeneca, initially as a medicinal chemist in the Infection, Cardiovascular & Metabolism Research Areas, then subsequently building technology platforms for Discovery and subsequently Global Process R&D. His roles at AstraZeneca have predicated upon external collaboration for successful project delivery.
Johanna (Hanneke) Jansen
Hanneke is a Director at the Novartis Institutes for BioMedical Research and heads the Computer-Aided Drug Discovery (CADD) group at their Emeryville campus. Her group develops and applies CADD approaches to projects at all stages of the global portfolio to impact drug discovery. Hanneke’s research interests include the design of relevant compound sets for hit-generation, improved predictive modeling that leverages large complex datasets, and developing computational chemistry & data-mining methods that can be integrated into state-of-the-art workflows to inform decision-making from target-ID and validation through hit generation and optimization. Prior to Novartis, Hanneke worked at Chiron Corporation and at Astra.
Hanneke is a Fellow of the American Chemical Society and the founder of the Teach-Discover-Treat initiative, which is an initiative to provide high quality computational chemistry tutorials that impact education and drug discovery for neglected diseases. She received her Ph.D. in Computational Medicinal Chemistry from the School of Pharmacy, University of Groningen (Netherlands) and completed a postdoctoral term at the BioMedical Center of Uppsala University (Sweden).
Adam Kallel
Adam brings over 25 years of computational chemistry experience to Retrophin. He received his Ph.D. in theoretical organic chemistry from UCLA, working with noted theoretical organic chemist Kendall N. Houk. He has authored multiple patents, 10 at Vertex Pharmaceuticals, with more being written. Adam has also supported many basic research programs that have led to pre-clinical and clinical programs. At Ligand Pharmaceuticals he led the Thromcytopenia program leading to LGD4665(Totrombopag). His expertise includes drug discovery in women’s health, anti-infective, metabolic diseases, neurological diseases, oncology and hematopoietic disorders. Adam is also considered to be an expert in the statistical analysis and graphical display of biological data.
Sylvie Sciammetta
Following a PhD in organic chemistry at the University of Leeds in the UK, Sylvie took a postdoctoral research fellowship at the University of Geneva in Switzerland. In 2000 Sylvie joined BioFocus (now part of Charles River Laboratories) where she led and advanced multiple medicinal chemistry projects on GPCRs to successful collaboration milestones. Sylvie joined Cresset in 2017 where, as an application scientist, she provides scientific and technical support to customers in North America.
Matthew Segall
Matt is CEO of Optibrium. He has a Master of Science in computation from the University of Oxford and a Ph.D. in theoretical physics from the University of Cambridge. As Associate Director at Camitro (UK), ArQule Inc. and then Inpharmatica, he led a team developing predictive ADME models and state-of-the-art intuitive decision-support and visualization tools for drug discovery. In January 2006, he became responsible for management of Inpharmatica’s ADME business, including experimental ADME services and the StarDrop software platform. Following acquisition of Inpharmatica, Matt became Senior Director responsible for BioFocus DPI’s ADMET division and in 2009 led a management buyout of the StarDrop business to found Optibrium, which develops software for small molecule design, optimisation and data analysis. Matt has published over 30 peer-reviewed papers and book chapters on computational chemistry, cheminformatics and drug discovery.
Tom Whitehead
Tom joined Intellegens straight from his PhD in theoretical physics at the University of Cambridge, and is now leading the application of Intellegens’ novel deep learning approaches to a wide variety of industrial applications. Tom is interested in developing machine learning approaches to solve previously intractable problems, in drug discovery and elsewhere.
Registration is now closed
The Alexandria, 10996 Torreyana Road, San Diego, CA 92121
Phone: 858.260.5336