Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
Libraries of chemical compounds are the lifeblood of modern drug discovery programs. The quality of library design can determine a project’s success or failure.
Both molecular modeling and cheminformatics techniques are important for the production of chemical libraries. The Cresset Consulting Services team has the analysis and design experience that is vital for the delivery of successful chemical libraries.
Library design as a concept is not new, but it only became a popular paradigm in drug discovery a decade or so ago. Over time the field of library design has split to encompass two main type of library, both of which are commonly used by medicinal chemists for their drug discovery campaigns:
Modern drug discovery now rarely proceeds simply via the classical route of making serial changes and acting on the output of testing. Rather, activity is explored using SAR explosions at discrete points in the process.
Diverse sets of compounds – be they drug-sized, lead-like or fragments – are usually created by selecting compounds from a greater pool using some measure of diversity on the pool. The pool could be commercially available compounds (singles or libraries), internal collections or synthetically accessible library space. Often a combination of these sources is used to get the widest possible range of compounds into the final library. In most cases the selection of compounds to include in the diverse library proceeds by using a combination of 2D similarity matrices and property calculations. This is essentially the process used by big pharma to get the most out of their compound screening file.
Although there are established methods for this, which work OK for generic screening molecules from vendors, there is no standard protocol and each company may have a different preferred set derived from the same commercially available pool.
With the rise of fragment based drug discovery over the last 5-10 years a thirst has emerged for libraries containing smaller lead-like and fragment-like diversity. The type of analysis required to gauge redundancy in this case becomes tricky as the smaller the molecules become the more difficult it is to create meaningful robust measures of chemical similarity – many of the 2D similarity methods lose their discriminatory ability. Thus fragment libraries or lead-like libraries may require special treatment.
We have become interested in using our own description of molecules – their shape and electrostatic character – to describe compound collections. We presented some initial work in this space at the spring 2012 ACS meeting. In this blog post Tim describes how we are looking again at the diversity of compound and specifically fragment collections using the computational efficiency available from BlazeGPU.
To design a focussed library computational input becomes a critical factor. Focused libraries are inherently the result of leveraging the designs using existing knowledge. However this knowledge can be applied in different ways. Two clear approaches are common in this space, each with differing factors that dictate the course of the library design workflow.
The technique typically used by compound vendors is to filter their compound collection based on the fit of molecules to activity models that have been developed (e.g. using physical property, pharmacophore or 2D similarity models). The usefulness of the classification is entirely dependent on the details of how the model has been constructed and applied.
The alternative technique, often employed by specialist vendors and bigger drug discovery organisations, is to design novel scaffolds and substitutions to address specific biological target areas of interest. These include application of structure or ligand based designs targeting protein families or sets of related targets using medicinal chemistry principles. Unlike the filtering approach above, in this case all molecules would have to be synthesized with inherent advantages (notably IP) and disadvantages (cost) that comes with this.
The latter undoubtedly requires the greatest engagement of time and resource to provide a suitable level of insight into the problem from which to develop innovative chemical solutions.
S-adenosyl methionine (SAM) is a co-factor used as a biological methylation synthon. It is employed in a host of enzymatic methyl transferase processes which are important in a number of disease areas. In the area of Epigenetics the lysine methyl transferases ‘KMT’s are responsible for methylating lysine groups on histones – a process which mediates gene expression by changing the stability of the nucleosome.
A quick analysis of the binding conformation of SAM across the PDB (Figure 1) reveals a small number of clusters of SAM bioactive conformations are observed. The conformation of SAM found in KMT’s form a tight cluster which is distinct from the more diverse generic SAM utilising enzymes. Interestingly, the analysis shows that DOT1L, which is also thought to be a KMT, is an outlier and more closely related to the generic enzyme set than to the other KMTs.
Assuming we wished to pursue a SAM mimetic design as a paradigm for KMT or DOT1L inhibitor generation, then from a molecular design point of view there are a number of issues which would need to be addressed. One major issue already given is that SAM is ubiquitously used as a cofactor thus a close mimetic may have unwanted side interactions. Clearly a DOT1L SAM mimetic design will have more issues with generic SAM enzyme crossover. A design aimed at other KMTs (e.g. SMYD2) would have selectivity issues just within the specific KMT family.
Designing away from potential crossover activity could be achieved by a full SAM mimetic design since both the adenine and Met chains adopt different vectors and shapes in the different sub-classes. Alternatively, concentrating on the adenine mimetic alone, the H-bonding patterns and solvent exposure are distinct in the two enzyme sub-classes as shown in Figure 2.
This simple example shows how some background knowledge on the system can impact on the scope and potential success of any given design.
We described in our previous blog how our fragment replacement tools can be used to search for novel bioisosteric replacements – in this case using the Spark software with adenine as the molecular input you can find suitable replacements as seeds for a library. As the template is extracted from a protein context all the ideas would be generated in the same coordinate frame and thus could be visualized and assessed for fit into the protein.
Alternatively the whole SAM 3D conformation from whichever sub-class could be submitted to Blaze to search for commercial vendor molecules that fit specific field patterns from the specific SAM conformation.
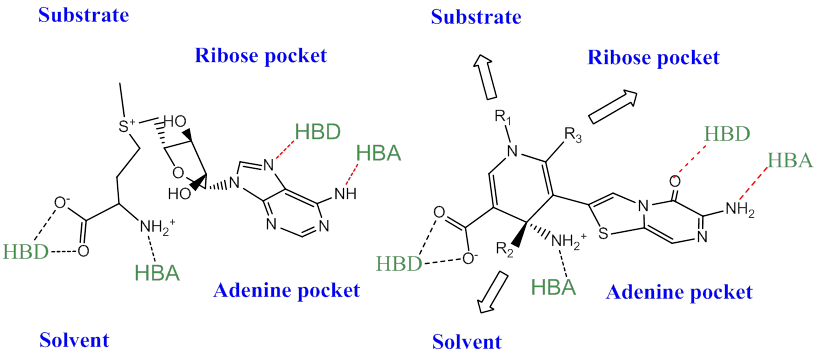
Figure 3. Library design idea for a SMYD-like KMT inhibitor (Left: SAM from SMYD2 and Right: virtual molecule)
The output of these virtual exercises, rather than being molecules to test (which is the usual scenario) would be molecular scaffolding ideas that would be potential starting seeds for a design. Ideally we would be looking for a good molecular fit to the interaction patterns (Figure 3) and especially to those which also provide appropriate synthetic vectors from which to explore the allowed variation defined from the starting binding pose.
In this case Spark has provided us with a design idea which matches well to the field patterns and interaction patterns required by the KMT SAM conformation in SMYD2 (PDB: 3S7F) and provides three potential vectors for a library: R1 for the substrate pocket, R2 for the open solvated pocket, R3 for the ribose pocket (Figs 3 and 4).
A standard protocol for constructing the library might proceed as follows:
Cresset computational chemists have wide knowledge of and experience in delivering projects involving all of the library scenarios described above which we are now able to offer as a service. Contact us for more information.