Future-proofing Cybersecurity in Drug Discovery
The pharmaceutical and biotech sectors suffer more data security breaches than any other industry, with 53% resulting from malicious activity. To protect against potential ...
News
In September I attended the RDKit User Group Meeting, which this year was (very conveniently for Cresset) held in Cambridge, UK, kindly hosted by Andreas Bender at the Department of Chemistry.
As most people will already know, the RDKit is an open-source cheminformatics toolkit developed by Greg Landrum, with regular and ongoing contributions from the community through its GitHub page.
As an RDKit enthusiast, the RDKit UGM is one of my favorite scientific events in the year. It is characterized by a relaxed and informal atmosphere: people attend to show what they achieved through the RDKit, or what they contributed to it, and to learn from others.
We have been building the RDKit into Cresset tools for a few years now, and have also contributed code to the project.
This year I gave a lightning talk about the integration of the RDKit in Flare, our structure-based design tool, and in particular on its usage from Python through the built-in Jupyter Notebook interface. Last month I wrote the blog post Integrating Jupyter Notebook into Flare on this topic, which you may find of interest. Actually, you’d better not get me started on the Flare Python Notebook or I might go on ranting for a long time, as I am terribly excited by the possibilities it opens up, and by how easy it makes it to implement one’s ideas and workflows!
But for now I’ll make an effort to stick to the topic of this post, which is a review of highlights of mine from the last RDKit UGM. As usual, the list is far from exhaustive – there were far more interesting talks than can fit in a blog post!
Greg Landrum (KNIME) started the meeting with the usual overview of the new features introduced since the last UGM. The C++ codebase has been largely updated to C++14, with a 20-30% increment in performance, which is quite impressive. The ETKDG conformer generator developed in 2015 by Greg and Sereina Riniker (ETH Zurich) has now become the default, and has been proved to be on par with commercial alternatives by two independent literature reviews [1, 2]. Further new features of the 2018.09 release include a new JSON-like chemical interchange format, CoordGen-based 2D depictions, and the possibility to instantiate RDKit molecules from SVG depictions endowed with chemical metadata. Finally, a nifty tool written by Nadine Schneider (Novartis Institutes for BioMedical Research) for CheTo to visualize fingerprint bits has been incorporated in the RDKit (Figure 1).
Figure 1. Fingerprint bit visualization.
Susan Leung (PhD student, University of Oxford) presented the 3-month Google Summer of Code project that she has been working on, which consisted in porting Matt Swain’s (Vernalis) MolVS, which had originally been written in Python, to the main C++ codebase, while expanding its current capabilities. MolVS is a great tool for molecule validation and standardization, i.e., all those automated operations to carry out when importing a set of chemical structures into a cheminformatics toolkit to get them in a consistent and manageable state (Figure 2).
Susan did a great job, even if the tautomer enumeration/canonicalization still has to be completed. This work is going to be very useful to the community and will ensure wider adoption of consistent molecular standardization criteria.
Figure 2. Some of the operations that MolVS can carry out on a chemical dataset.
Pat Walters (Relay Therapeutics) presented three programs built on top of the RDKit and available through Pat’s GitHub page.
The first program addresses the case, quite common in pharmaceutical companies, where hundreds of compounds have been synthesized as part of a project, but still these compounds do not cover all possibilities: there are certainly holes and potentially interesting molecules that were missed. In such cases, a Free-Wilson analysis allows to evaluate the contributions of different R-groups and identify candidates to prioritize for synthesis. Molecules are decomposed into R-groups, a matrix which encodes presence or absence of each R group in each molecule is generated, then R-groups vector are regressed against pIC50 values. Linear regression does not usually work well for this purpose, as many characteristics are collinear, so Ridge Regression is a better choice to avoid hitting the collinearity problem.
The second program is aimed at filtering chemical libraries via the functional group filters from the ChEMBL database, and some property filters from the RDKit, which act as structural alerts to identify potentially problematic molecules at an early stage.
The last program is an implementation of Delaney’s ESOL method to predict aqueous solubility from molecular structure. When discussing the results, Pat showed a chart comparing the r2 obtained by his ESOL implementation to those obtained through RF, DNN and Graph Convolution methods.
While apparently ESOL gets a higher r2, the difference is not statistically significant, because of the confidence intervals associated to the number of data points (Figure 3). This is a point that Pat has already stressed in his blog post Predicting Aqueous Solubility – It’s Harder Than It Looks, and certainly worth keeping in mind.
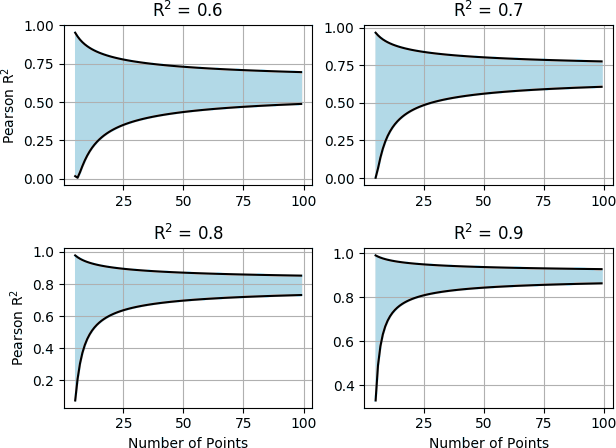
Figure 3. The light blue area shows the 95% confidence interval for r2 depending on the number of data points.
Ben Tehan and Rob Smith (Heptares) presented an overview of how the RDKit is used at Heptares. In an effort to harmonize and simplify their cheminformatics infrastructure, Heptares adopted the RDKit as their standard platform in 2013. I was pleased to learn that in their hands the performance of my Open3DALIGN alignment algorithm (which indeed was my first contribution to the RDKit, back in 2013) coupled with shape-based scoring was not too far from ROCS (EF1% ~10 using CrippenO3A descriptors compared to ~15 enrichment with ROCS). So I gave myself a pat on the back and carried on.
For those who have never been to an RDKit UGM, I’ll do a small preamble. Traditionally, Roger Sayle (NextMove) gives a talk where he picks some RDKit algorithm, pinpoints how poorly coded it is in its current implementation, and how it could be written much better. This is usually quite enjoyable (Greg might have a different opinion), particularly because the talk is always accompanied by a GitHub pull request by Roger with the improved code, to the benefit of the community.
This year the focus was a bit different: rather than tearing some RDKit bit into pieces, Roger decided to show the potential performance pitfalls behind apparently simple common (chem)informatics tasks, such as counting text lines in a file or computing molecular weight, to finish by spotting out a claim in a scientific paper based on a wrong percentage calculation. While very interesting as usual, I thought that the arguments in favor of low-level programming to speed up certain tasks were not entirely convincing, as they were not supported by timings showing that the extent of the gain actually balances the losses in terms of effort and portability.
Once again, this was a thoroughly enjoyable meeting. If you’re interested in finding out more, see the materials from the RDKit UGM 2018.