Discovery CRO
Prioritization of new molecule design using QSAR models - 2D- and 3D-QSAR studies on SARS-CoV-2 Mpro inhibitors
Introduction
The viral main protease Mpro is a crucial enzyme for the replication of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of the COVID-19 global pandemic. In addition to the established vaccination programs against COVID-19, antiviral drugs are seen as essential to control the inevitable future epidemics of coronaviruses. Because of its key role, Mpro has received much attention as a potential target for novel therapeutic agents.1-6
Robust and predictive Quantitative Structure Activity Relationships (QSAR) models of activity against the Mpro of SARS-CoV-2 can be developed to elucidate observed activity and inform new molecule design. The Cresset Discovery CRO team used a dataset of 76 compounds with known experimental activity and common binding mode1-6 to build both predictive machine learning (ML) and the Field 3D-QSAR methods in the molecular modeling solution FlareTM.7
All models showed statistical performance aligned with experimental results, and with comparable descriptive and predictive abilities across all models. Additionally, the Field 3D-QSAR method highlighted molecular regions driving activity.
Methods
Dataset
For this case study, a dataset of 76 non-covalent inhibitors with different chemotypes and an evenly distributed activity range (pIC50: 4.00 – 7.74) was partitioned into training set (56 molecules) and test set (20 molecules) using a 26% activity stratification.
2D-QSAR
The initially selected 2D descriptors (some imported from RDKit8) were cross-correlated by a linear Pearson correlation matrix to reduce redundant descriptors. Using 6 physico-chemical descriptors (MW, TPSA, #RB, NumHAcceptors, NumHDonors and RingCount) and fingerprints (RDKit, Morgan and MACCS keys), 2D-QSAR regression models were developed using the supervised machine learning (ML) methods; Support Vector Machine (SVM)9, Gaussian Process Regression (GPR)10, Random Forest (RF)11 and Multilayer Perceptron (MLP)12.
3D-QSAR
Building 3D-QSAR models is highly sensitive to conformation searching and molecular alignments. The high-quality alignments performed by Flare, in particular those based on the maximum common substructure (MCS) algorithm, combined with Cresset Discovery’s expertise to interpret the results, generated meaningful molecular alignments with a low degree of noise (Figure 1). The compounds were aligned by MCS to the co-crystallized ligands of the PDB IDs 7L131, 7L141, 7QBB5 and 8SXR6 which were used as references (weighted average contribution) and using a 'Soft' hardness of the 7L13 protein as an excluded volume. The conformation hunt parameters used were the standard 'very accurate and slow' (within a 2.5 kcal/mol energy window).
The 3D-QSAR regression models were developed using the Field 3D-QSAR method (standard 'Normal with Y scrambles' scheme) and the machine learning methods: k-Nearest Neighbors (kNN), SVM, GPR, RF, MLP and Consensus.7 These methods use probe positions directly determined from the field points attained from the Cresset XED force field to sample the electrostatic potential and volume/shape for each molecule in the training set, which can then be used as descriptors for the QSAR models. The exception was the kNN method which uses the Cresset XED force field as a metric of similarity.7
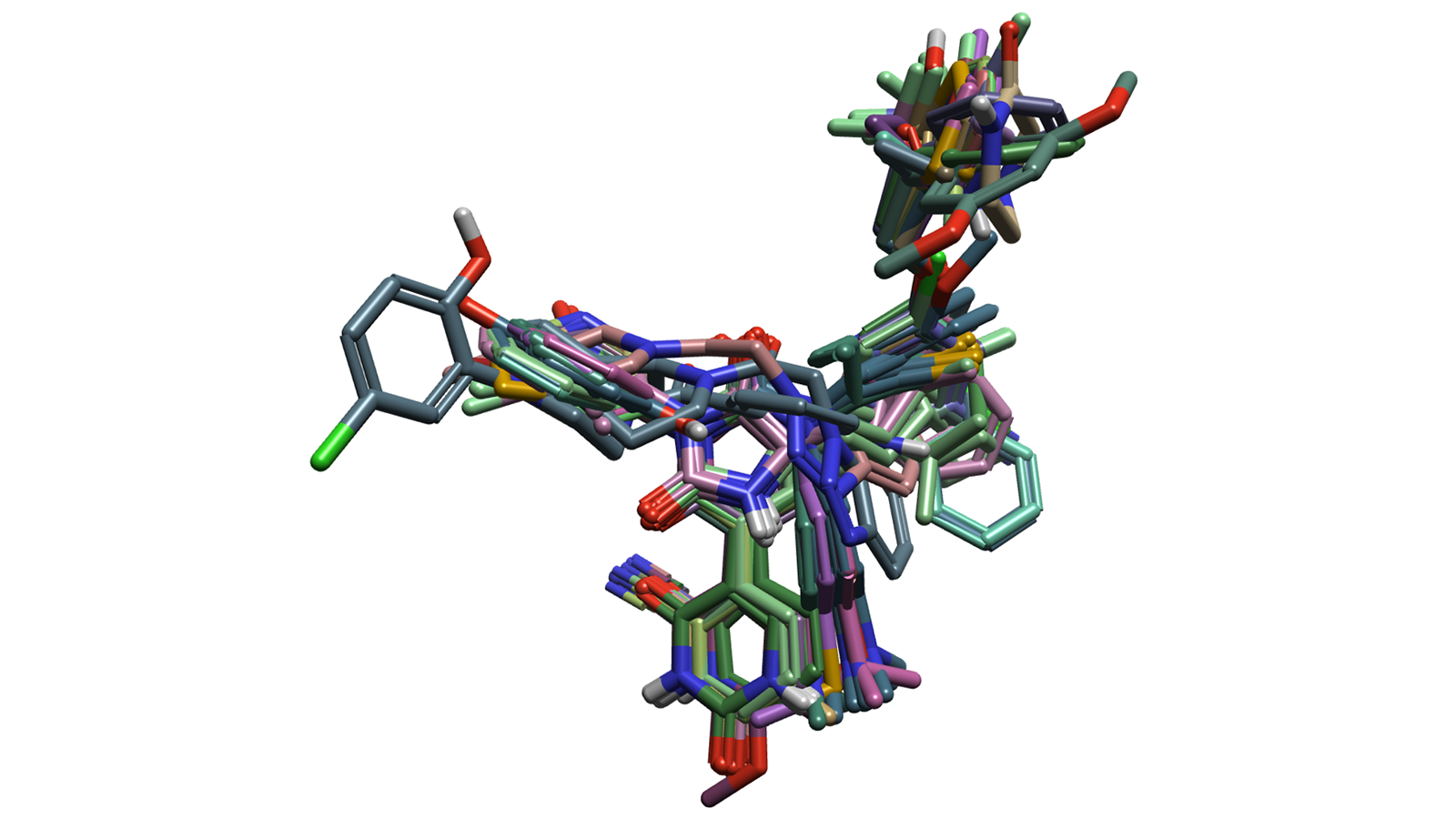
Figure 1. Representation of the molecular alignment of the dataset of compounds.
Results
Statistical analysis
Table 1 shows the performance of each model, ranked by the prediction statistics for all assessed QSAR methods. Figure 2 shows the experimental vs. predicted activity plots for both Morgan FP MLP 2D-QSAR and the MLP 3D-QSAR models which have the highest overall r2 test set of 0.72. The confidence in the models, as determined by the r2 of all test sets, is high and comparable with the exception being the MACCS keys SVM 2D QSAR. Hence, any of these models would provide an accurate prediction of the activity of new compounds. The good agreement between the 2D and 3D models suggests that the compounds of this dataset act via a similar mechanism. It also proposes the RDKit 2D descriptors and fingerprints as good alternatives to the use of Cresset 3D descriptors for building predictive ML models. However, contrarily to the Cresset molecular field points, these models do not provide insights to potential key regions for further modeling in order to improve the recognition, binding and subsequently the activity of the tested molecule.
Table 1. Comparison of the different QSAR model measured and predicted statistics.
| QSAR type | Regression model | r2 training set | q2 training set CV | r2 test set |
|
2D-QSAR
|
MLP | 0.91 | 0.68 | 0.69 |
| GPR | 0.89 | 0.73 | 0.67 | |
| Consensus | 0.89 | 0.74 | 0.65 | |
| RF | 0.86 | 0.74 | 0.62 | |
| SVM | 0.86 | 0.75 | 0.61 | |
|
2D-QSAR
|
MLP (Morgan FP) | 1.00 | 0.80 | 0.72 |
| SVM (RDKit FP) | 1.00 | 0.83 | 0.63 | |
| SVM (MACCS keys) | 0.96 | 0.80 | 0.50 | |
| 3D-QSAR | MLP | 1.00 | 0.82 | 0.72 |
| Field QSAR | 0.96 | 0.81 | 0.71 | |
| kNN | - | 0.75 | 0.71 | |
| Consensus | 0.99 | 0.82 | 0.70 | |
| SVM | 0.98 | 0.82 | 0.70 | |
| GPR | 0.99 | 0.77 | 0.70 | |
| RF | 0.99 | 0.82 | 0.70 |
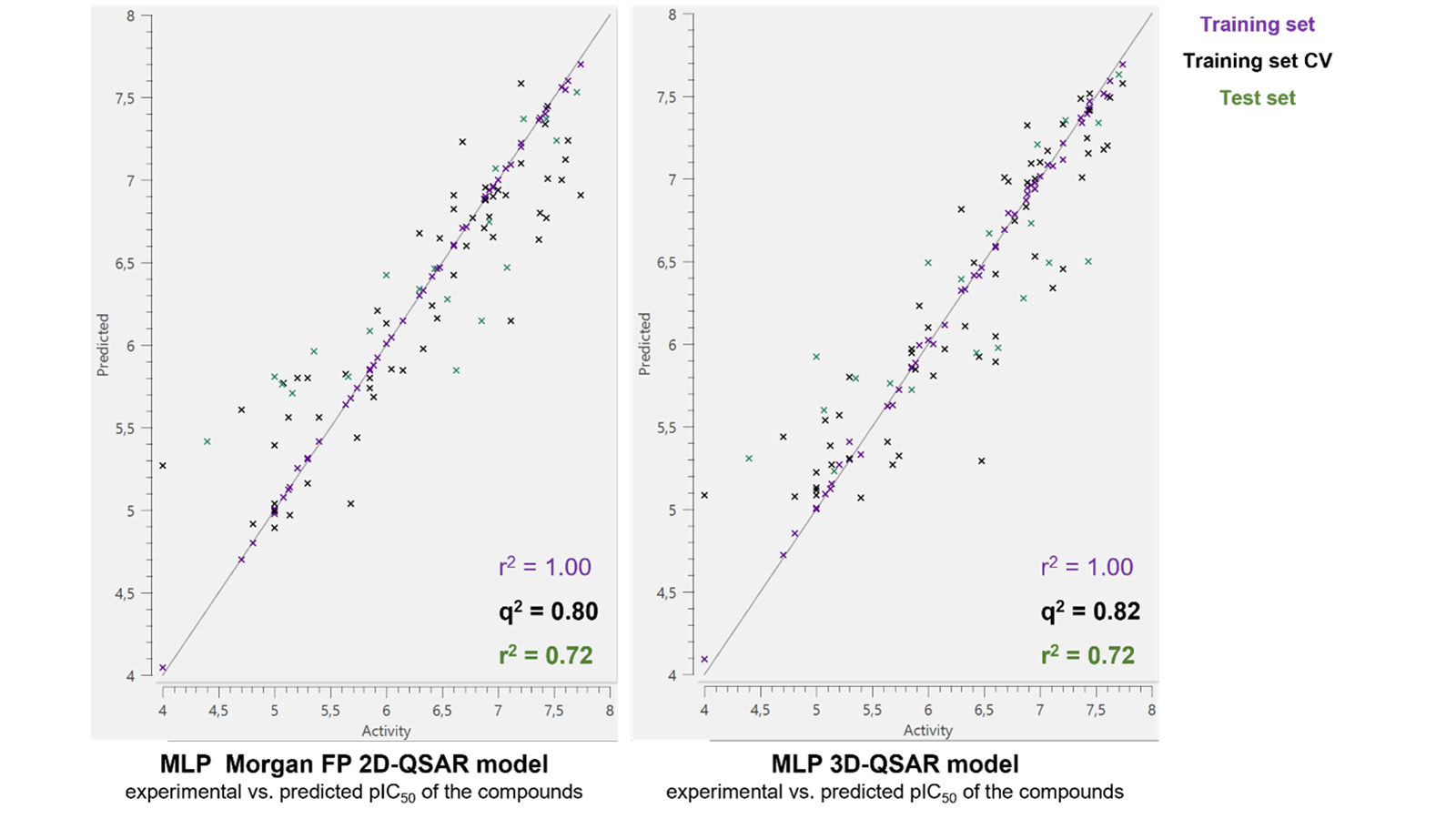
Figure 2. MLP Morgan FP 2D-QSAR (left) and 3D-QSAR (right) models - experimental vs. predicted activity of the compounds in the training set (purple), training set Cross Validation (CV) (black) and the test set (green).
Model visualization and interpretation
The unique Cresset Field 3D-QSAR method offers the advantage over ML methods in that the visual inspection of the model coefficients identifies regions where the model predicts strong effects on activity. Figure 3 illustrates the electrostatic and steric model coefficients superposed to the most potent molecule (37, pIC50 = 7.74). Regions of favorable negative electrostatic coefficients are observed in the amide-carbonyl of the core ring and the nitrogen atom of the pyridine unit, which implies that a less positive charge on these regions improves activity. Additionally, the large green dots point out regions of favorable steric coefficients near the 2-chlorobenzyl moiety, which in combination to the high steric variance verified, suggest this is the best moiety to model (to increase potency). Using the learning from the model and the computational and medicinal chemistry know-how of Cresset Discovery's team, proposals of inclusion of cyano or methyl substituents at the 3rd position of the 2-chlorobenzyl unit result in improved activity, with superior potency than those predicted for compound 37 (predicted pIC50 of 7.70 and 7.60, respectively). In addition to the steric effects, these groups optimize the intermolecular interactions with the enzyme (CN allows a hydrogen bond with the backbone-NH of Q192, whilst the methyl makes hydrophobic contacts with P168 and the alkyl region of the Q192 side chain) that subsequently could improve the inhibitor potency.
Furthermore, the relevance of the 2-chlorobenzyl alcohol group is highlighted by comparing the field contributions of compound 37 with similar molecules (Figure 4). The absence of this group in compound 8 has an unfavorable electrostatic contribution that accounts in a decrease in activity by ca. 2.5 log units; whilst large, and mild unfavorable electrostatic and steric contributions are observed with the substitution of the aromatic ring by an alkyl chain or a saturated ring, causing a decrease in activity of ca.1 log unit. Similarly, the presence of a hydroxyl group such as in compound 28 has a strong unfavorable electrostatic contribution which decreases its predicted activity.
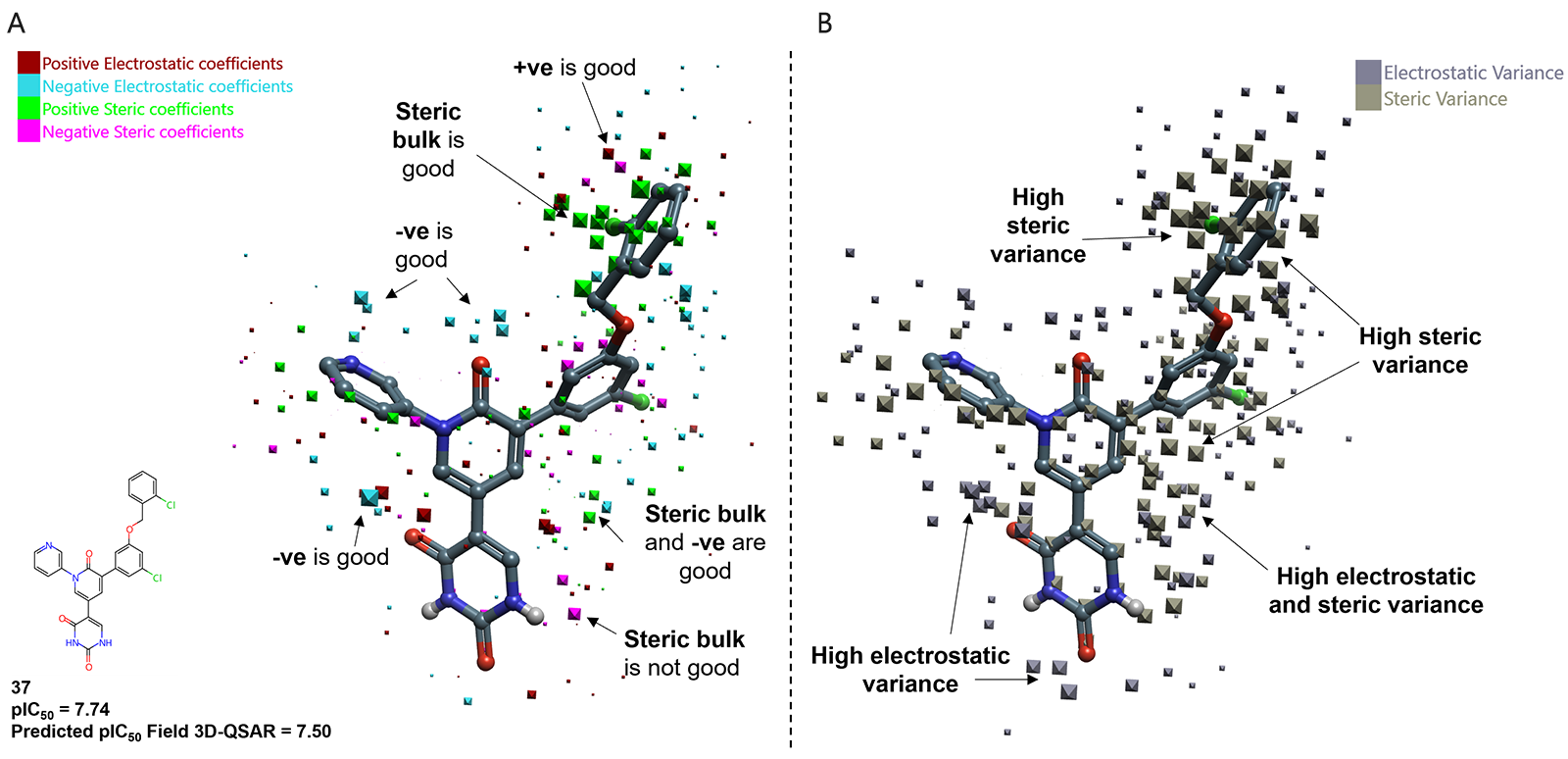
Figure 3. Model coefficient plot for the SARS-CoV-2 Mpro Field QSAR model. (A) electrostatic and steric coefficients; (B) electrostatic and steric variance, using the most potent molecule (37) as reference. Compounds numbering is according to the patent WO2022/150584A1.4
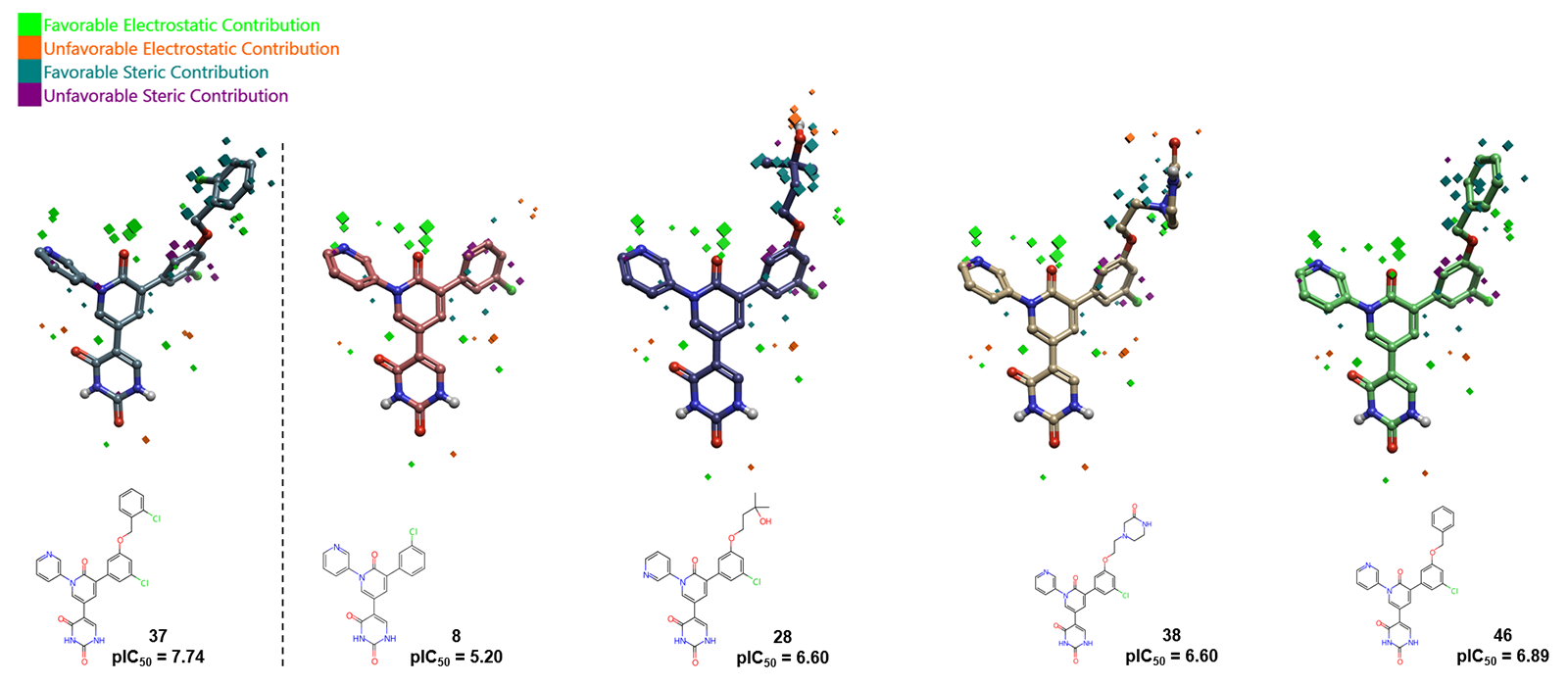
Figure 4. SARS-CoV-2 Mpro 3D-QSAR field contributions to predicted activity for compounds 37, 8, 28, 38 and 46.
Conclusions
Cresset Discovery's scientists successfully built robust 2D-QSAR and 3D-QSAR regression models to describe and predict the activity of a library of non-covalent SARS-CoV-2 Mpro inhibitors. We illustrate the agreement between 2D- and 3D-QSAR models and the superior performance of the Field 3D-QSAR model over the machine learning methods. The team's expertise further enabled the analysis of the electrostatic and steric coefficients to rationalize inhibitor potency and highlighting how to guide and prioritize the design of novel therapeutic molecules.
The robust QSAR methods within Flare, combined with Cresset Discovery CRO knowledge in quantitative structure-activity relationship methods provides a premium service to accelerate the design of innovative drugs. Reach out to learn how Cresset Discovery CRO team can support, clarify, or even carry out your QSAR projects.
References
- Chun-Hui Zhang, et al., ACS Cent. Sci. 2021, 7, 467–475, https://doi.org/10.1021/acscentsci.1c00039
- Chun-Hui Zhang, et al., ACS Med. Chem. Lett. 2021, 12, 1325–1332, https://doi.org/10.1021/acsmedchemlett.1c00326
- Maya G. Deshmukh, et al., Structure 2021, 29, 823–833, https://doi.org/10.1016/j.str.2021.06.002
- William L. Jorgensen, Patent WO 2022/150584 A1
- Andreas Luttens et. al., J. Am. Chem. Soc. 2022, 144, 2905–2920, https://doi.org/10.1021/jacs.1c08402
- Jimena Perez-Vargas et. al., Emerg. Microbes Infect. 2023, 12, 2246594, doi.10.1080/22221751.2023.2246594
- Flare™, Cresset®, Litlington, Cambridgeshire, UK; https://www.cresset-group.com/software/flare/; Cheeseright T., Mackey M., Rose S., Vinter, A.; Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation J. Chem. Inf. Model. 2006, 46 (2), 665-676.
- RDKit: Open-source cheminformatics. https://www.rdkit.org
- Harris Drucker et. al., Support Vector Regression Machines, Advanced in Neural Information Processing Systems 9, 1996, 155-161.
- C. E. Rasmussen, C. K. I. Williams, Gaussian processes for machine learning 2016, The MIT Press, ISBN 026218253X.
- L. Breiman, Random Forests, Machine Learning, 2001, 45, 5-32.
- F. Rosenblatt, Principles of Neurodynamics: Perceptrons and the Theory of Brain Mechanisms, Washington: Spartan Books, 1962.
Contact us for a free confidential discussion
We help you reach your next milestone faster and more cost effectively
Contact us for a free confidential discussion