Spark™
Scaffold hopping and R-group replacement
Quickly advance your drug discovery projects by finding novel and diverse biologically relevant replacements for both core and terminal groups
Find biologically relevant replacements to known active molecules in just a few simple clicks
Thanks to Spark’s ‘product-centric’ approach to bioisosteric replacement, users can generate more diverse, less obvious bioisosteres than other comparable methods.
- Specify the part of the molecule to replace
- Choose the chemistry of the attachment points, specifying the atom types you are willing to accept in replacement fragments
- Indicate the fragment source from a large collection of databases
- Form the product structures by merging matching fragments with the correct angles and distances into the starting molecule
- Score the product structures against the starting molecule, based on their electrostatics and shape similarity
Easy analysis of Spark results
Once the calculation is completed, Spark will present results as a list of biologically relevant replacements ranked using Cresset’s unique XED force field model of electrostatics.
- The 'Standard results' table shows individual results, sorted by similarity score
- The 'Clustered results' table shows the results sorted into chemical families
- The 'Tile results' table shows a summary view of the results, specifically designed to support medicinal chemists in their analysis
Visually meaningful analysis for molecule triaging and prioritization is supported with property-based traffic light coloring of the results table.
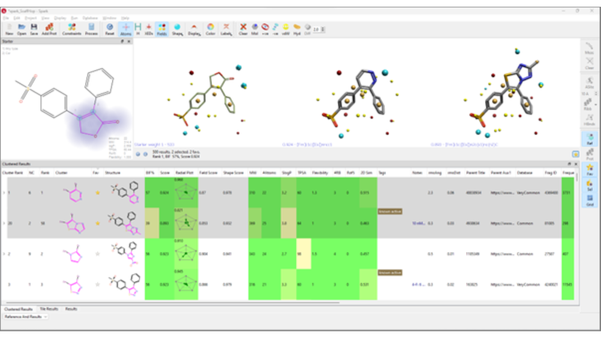
Selected Spark results with field points displayed, and color-coded properties to assist in compound profiling and candidate selection. For each molecule, the replacement fragment is highlighted in magenta, and displayed alongside additional information about the results.