The ‘Build Loops with FREAD’ extension is based on the FREAD structure prediction protocol developed by the University of Oxford1.
FREAD predicts loop structures by selecting likely conformations from a database of experimentally-determined protein fragments, starting from the assumption that loops with similar sequences also have similar conformations, taking into account the spatial constraints introduced by their anchor residues.
Download the FREAD loop database (these files are very large at 20GB).
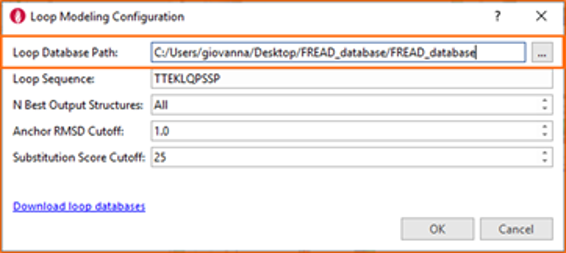
Once you have downloaded the .zip/.tar file, move it to the directory where you wish to install the database and extract its content. Within Flare, open ‘Build Loops with FREAD’, then set the ‘Loop Database Path’ option to point to the desired database location, as shown below:
References
- Choi, Y. and Deane, C. M. FREAD revisited: accurate loop structure prediction using a database search algorithm, Proteins (2009)