Kinase inhibitors have established themselves as cancer therapeutics but does this family of enzymes have a role to play in other areas of human disease? Prof. Michael Schneider, who heads the splendidly named ‘Cardiac Myogenesis, Death and Regeneration’ research group at Imperial College London, says that they do. He has long had an interest in the role that one such kinase, MAP4K4, plays in human cardiac muscle cell death. When we first met he had already established that this kinase was upregulated in diseased human heart tissue. Moreover, a series of carefully conceived biological experiments went on to suggest it played a pivotal role in cardiac muscle cell death- effects that could be recapitulated in cardiomyocytes derived from human induced pluripotent stem cells (iPSC-CMs). There is precedent for using these cells in drug discovery to screen drug candidates for adverse myopathic or proarrhythmic effects before they are trialled in humans, but this is the first published case of using them to screen molecules in the earliest stages of a drug discovery program. The barrier to developing new cardiac drugs is high and many agents have failed to demonstrate efficacy in the clinic. This has reduced the number of candidates coming to the market to a trickle – no drugs were approved for cardiac indications in 2018 against 13 for cancer.
In this case the Imperial group achieved a milestone by using human iPSC-CMs to screen novel MAP4K4 inhibitors for cardioprotection – effects that were later confirmed in vivo where they showed that inhibiting this kinase markedly reduced cardiac ischemia-reperfusion injury in mice. The discovery of the small molecule inhibitors and their role in protecting cardiomyocytes from damage recently appeared in the April 2019 edition of Cell Stem Cell.1
The challenge: Find a chemical starting point
No MAP4K4 inhibitors had been described when Imperial’s Drug Discovery Centre took up the challenge of finding an initial tool compound. Similarly, the absence of any protein structure ruled out using a docking application to run a virtual screen. We developed a three-stage workflow to find chemical tools we could use to interrogate MAP4K4 biology and Cresset software played a central role in this project.
Solution
Step 1
In practise, most kinase inhibitors target more than one enzyme and we decided to screen a commercial set of ~1800 well characterized, biologically active compounds against the catalytic domain of MAP4K4. We identified a small set of compounds – all known kinase inhibitors, with significant potency for MAP4K4 albeit at a lower level than their primary activities.
Step 2
To pull out the features responsible for the off-target MAP4K4 activity shared by these compounds. Cresset’s FieldTemplater™, a component of Forge™, is the ideal tool to find common field point features within a set of molecules. The output is a field point pharmacophore made up of an individual conformation of each of the three molecules. The familiar pattern of H-bond donor-acceptor-donor common to most ATP-competitive kinase inhibitors is apparent, along with additional clusters of negative (Cluster A) and positive (Cluster B) field points.
Step 3
To use this template as the seed for a virtual screen using Blaze™ to look for other molecules that shared these features. This gave us a hitlist of commercially available compounds that we could purchase to kickstart this project. F1386-0303 (MAP4K4 pIC50 7.5) was one of the first compounds tested and Michael’s group were able to show that MAP4K4 inhibitors protect hiPSC-cardiomyocytes from oxidative stress at levels equivalent to that of MAP4K4 shRNA.
A full medicinal chemistry program increased the oral availability this series to a point where an improved compound, DMX-5804, was shown to protect the heart in acute myocardial infarction. This combination of innovative biology and chemistry demonstrates the potential of MAP4K4 inhibition as a new target for treating heart disease.
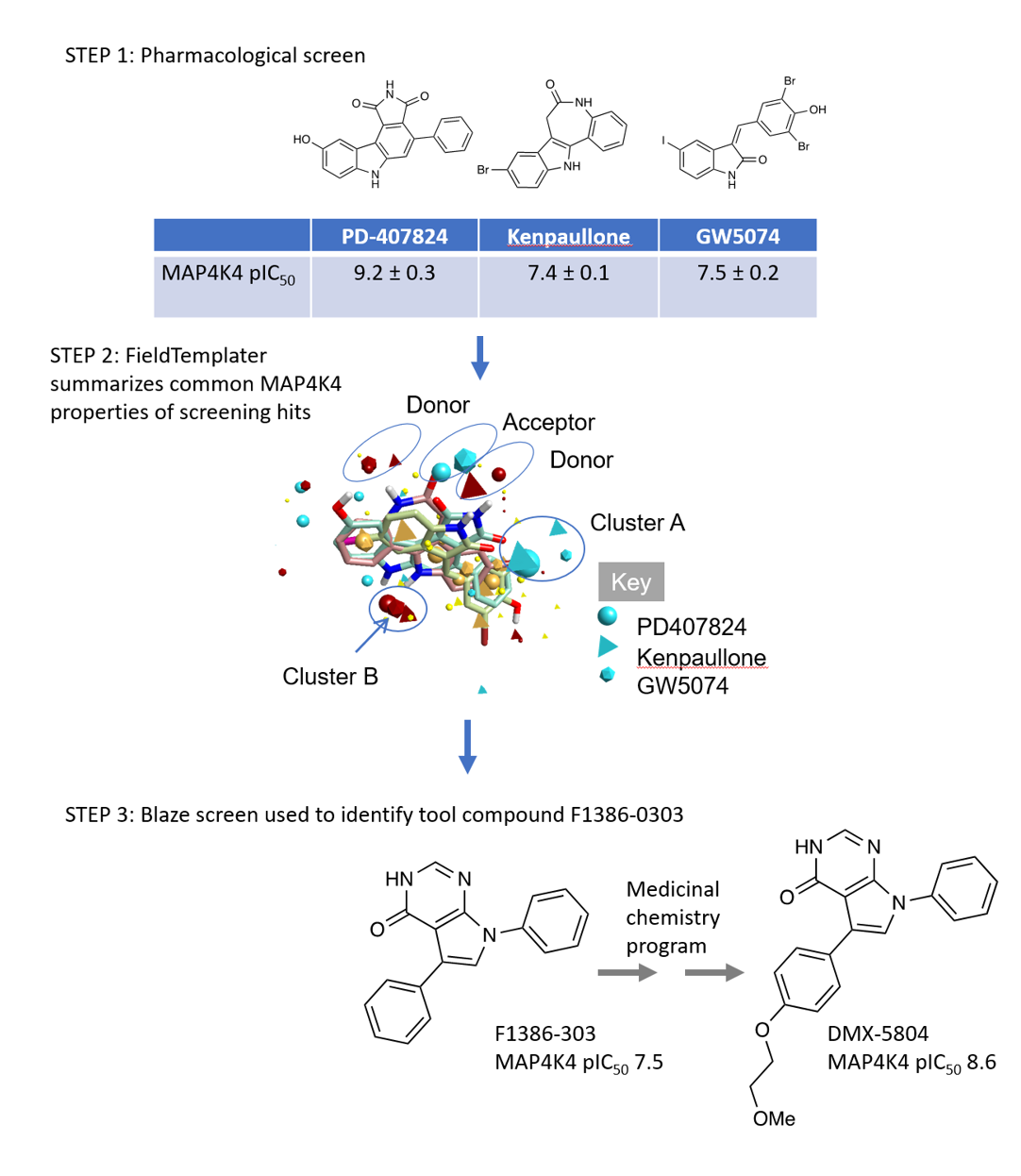
Three step workflow for generating initial MAP4K4 hit compound. STEP 1: pharmacological screen of small commercial library identifies MAP4K4 inhibition as an off target effect of 3 known kinase inhibitors. STEP 2: FieldTemplater extracts common features of the three molecules. STEP3: F1386-303 identified from Blaze screen run using FieldTemplater pharmacophore from STEP2. Activity and pharmacological properties improved following medicinal chemistry optimization to give DMX-5804.
Reference
- Fiedler, L. R. et al. MAP4K4 Inhibition Promotes Survival of Human Stem Cell-Derived Cardiomyocytes and Reduces Infarct Size In Vivo. Cell Stem Cell 24, 579-591.e12 (2019).
