Discovery
Fast-tracking drug repurposing research with virtual screening solutions
The Australian Program for Drug Repurposing for Treatment Resistant Ovarian Cancer is a five-year program conducting research into the repositioning of approved drugs to explore new therapeutics for treatment-resistant high grade serous (HGS) ovarian cancer.
Started in 2020, the program aims to screen a total of 20 drug targets over five years. The research team’s strategy has focused on the creation of an annual drug repurposing pipeline, combining multi-stage in silico and in vitro methods, through to economic pre-modelling and clinical trial concept development.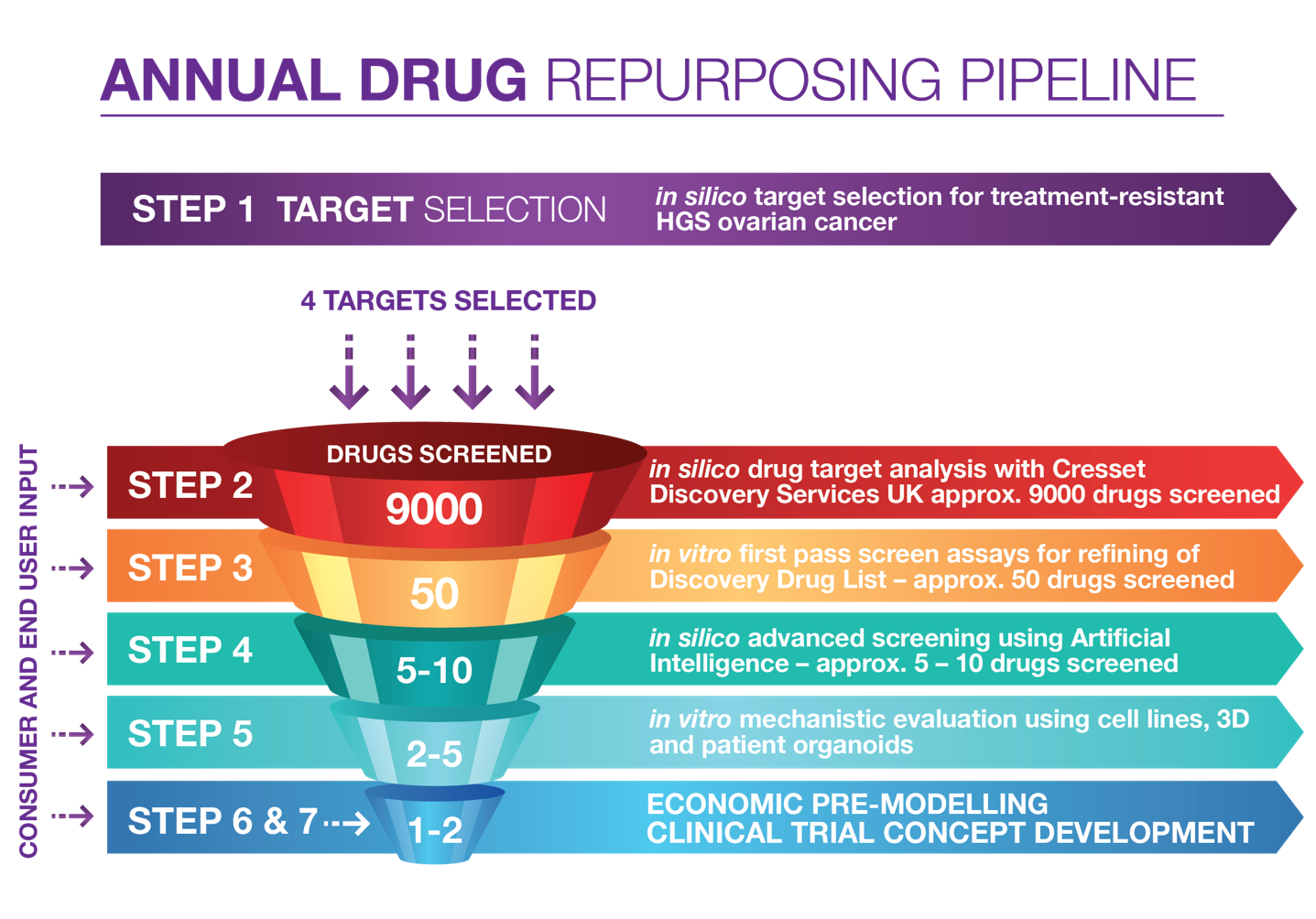
Figure 1. The Australian Program for Drug Repurposing for Treatment Resistant Ovarian Cancer – annual drug repurposing pipeline
High grade serous (HGS) ovarian cancer is the most common subtype of ovarian cancer and has a poor 5-year survival rate due to disease recurrence and resistance to current therapies. There is no early detection test, and in the absence of any specific symptoms, diagnosis usually occurs at an advanced stage. Patients with late-stage disease undertake an existing treatment program commencing with surgery, followed by multiple cycles of chemotherapy, until the patient relapses or develops new disease within six months of cycle completion, or becomes intolerant to the chemotherapy and can no longer receive the treatment. The spread and cellular complexity of the disease reduces the likelihood of a single therapeutic treatment being effective. Effective new therapies are therefore urgently needed.
In the initial step in the repurposing pipeline, potential druggable targets were identified using AI analysis of published literature, resulting in selection of four well-defined druggable targets specific to ovarian cancer.
At this stage, Cresset Discovery collaborated on the project to fully explore the four identified targets and their structures, and to use this data to carry out virtual screening to identify compounds with known safety profiles that had binding potential for the target.
The first stage involved analysis of the druggable target including sequence, structural biology and known structural ligands for that target, as well as exploring any strategically related target receptors with a similar structure. This data was used to investigate the structure of the protein and potential binding sites to determine a strategy for virtual screening. The analysis identified two potential binding sites and two templates were created for virtual screening.
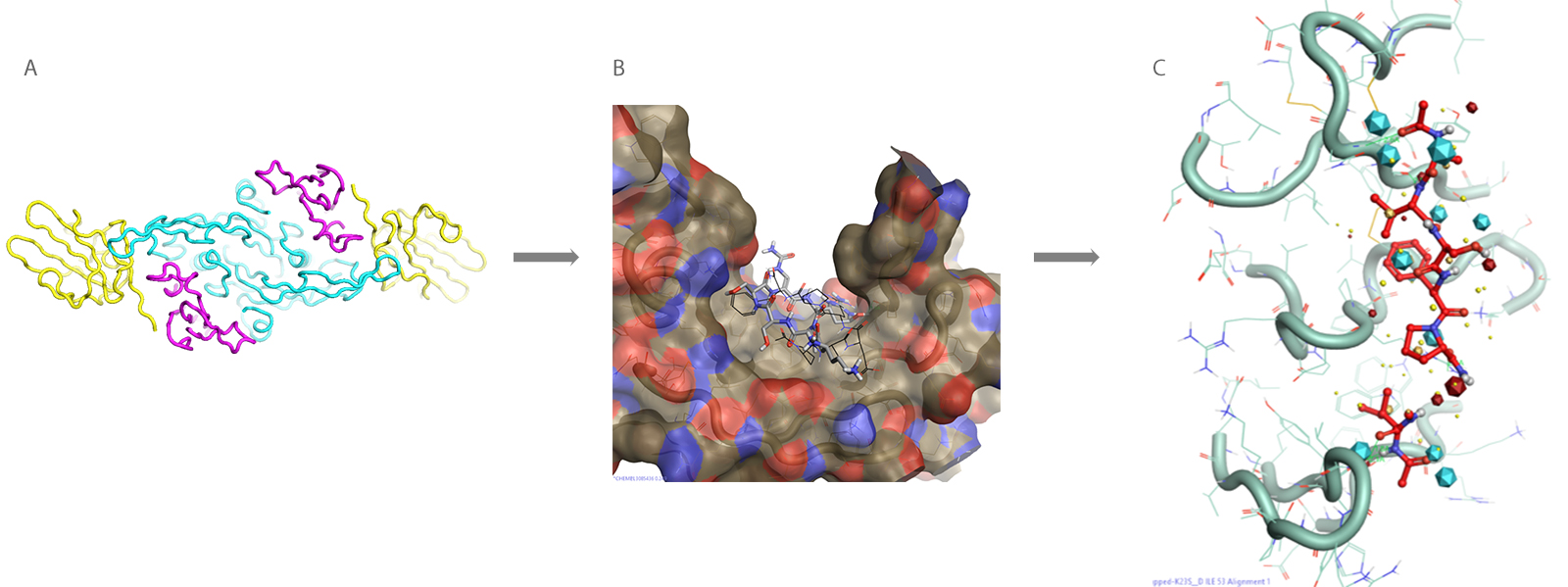
Figure 2. From analysis of the druggable target (A) to creation of the ligand-based template for virtual screening (B) to analysis of the electrostatic potential of the ligand protein interactions (C)
The two ligand-based templates were used to run virtual screens to search approximately 7500 drugs either approved by the Federal Drug Administration (FDA) or drugs going through clinical trials at the time of the screen1.
The virtual screen results were scored, and the hit list was analyzed further using multiple parameter optimization methods by Cresset’s expert team. The hits were docked and ranked2, and the protein-ligand electrostatic complementarity was calculated and scored3.
These three independent scores were combined to shortlist the final candidates for each of the two binding sites.
Visual inspection of each molecule was conducted by Cresset’s expert modelers to produce a final shortlist of plausible candidates that would bind to the target. Each candidate was then assessed for known oncology indication but having not previously been used to treat ovarian cancer.
The result was a list of approximately 50 drugs with potential interaction with the four targets, enabling prioritization for the next stage in the pipeline.
This list was provided to the Program team and further pharmacological filtering parameters were applied for final refinement, resulting in two drugs which showed potential for use in clinical practise combined with a high score for binding to the target identified by the virtual screen step by Cresset Discovery. These two candidates proceeded to in vitro analysis in the lab.
Drug A, an FDA-approved antiviral drug, was found to display anti-proliferative action, however the dose required was not achievable and the drug did not proceed through the pipeline. Drug B, another approved antiviral drug, was also shown to be effective and within an achievable dose, and this drug proceeded to undergo economic pre-modelling involving consultation with the program’s consumer advisory group and other stakeholders for development into a clinical trial concept.
Cresset Discovery combined their expertise in silico methods, insights from specialist solutions and their experience in medicinal chemistry to provide powerful computational drug target analysis, to prioritize therapeutics for further investigation.
Find out more about The Australian Program for Drug Repurposing for Treatment Resistant Ovarian Cancer and Cresset’s collaboration by watching our webinar with Professor Nikola Bowden of the University of Newcastle.
References
- Blaze, Cresset®, Litlington, Cambridgeshire, UK; https://www.cresset-group.com/software/blaze/
- Lead Finder™, BioMolTech®, Toronto, Ontario, Canada; https://www.cresset-group.com/software/lead-finder/
- Flare™, Cresset®, Litlington, Cambridgeshire, UK; https://www.cresset-group.com/software/flare/; Cheeseright T., Mackey M., Rose S., Vinter, A.; Electrostatic Complementarity as a Fast and Effective Tool to Optimize Binding and Selectivity of Protein–Ligand Complexes J. Med. Chem. 2019, 62, 6, 3036-3050;
Discovery CRO
Contact us for a free confidential discussion
We help you reach your next milestone faster and more cost effectively
Contact us for a free confidential discussion