Keverne Louison, Scott Midgley†
†Cresset, New Cambridge House, Bassingbourn Road, Litlington, Cambridgeshire, SG8 0SS, UK.
Introduction
Cannabinoid (CB) receptors are a class of G-protein coupled receptors (GPCRs) that continue to be an active area of research across several therapeutic areas. This includes selective targeting of the CB2 receptor which could be a potent analgesic, but without the psychoactive effects of CB1 binding, and so potentially reducing the usage of opioid based analgesics and the associated risks of addiction. While CB2's immunomodulatory characteristics and capabilities may aid in treating obesity.1,2
Virtual screening can provide a starting point in searching for new potential selective, hit molecules. Virtual screens can be conducted on crystallographic ligands without any preparatory modeling, however, by combining structure-based computational methods, structural biology and chemistry knowledge, with ligand-based computational methods, we can optimize the starting point for a virtual screen, identifying compounds which may have improved receptor subtype selectivity.
This study demonstrates how analysis of the starting point and correct triaging of a virtual screen can be used to identify, select and prioritize compounds. The compounds can then either be purchased from the commercial libraries that were used in the screening, passed onto chemists or medicinal chemists to synthesize, and subsequently to pharmacologists for biological testing.
Method
In this study, we will begin with obtaining the publicly available crystal structures of both the CB1 and CB2 receptors. Analysis of the protein-ligand complexes with the aim to identify opportunities to selectively target CB2 ligands can then be undertaken.
Cryo-EM structures 6KPC (CB2) and 6KPG (CB1), and their corresponding, pre-clinical ligands (Figure 1), were downloaded from the Protein Data Bank (PDB) and prepared using Flare™.3,4 The G-protein coupled receptor (GPCR) domain of both protein structures were then aligned to clearly visualize the binding pockets of both receptors. This way, potential subtype specific residues and regions within the CB2 receptor could be identified to elicit target selective interactions with a new ligand. The native ligands for each receptor PDB structure were docked in the opposing receptor PDB structure to generate binding poses for the ligands in both receptors.
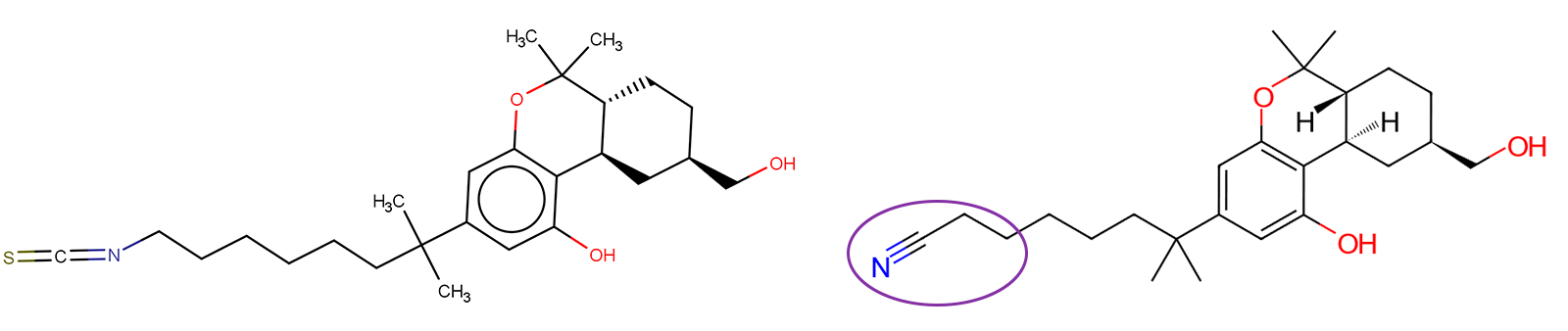
Figure 1. Cryo-EM ligands for CB1, AM12033 (PDBID: 6KPG)4 (left) and CB2, AM841 (PDBID: 6KPC)4 (right).
The highlighted atoms of the CB2 cryo-EM ligand AM841, circled in purple, (Figure 1) were selected to initiate the assessment of the receptor binding pockets. Conducting R-group replacement could help identify new structures where this pocket could be exploited by eliciting an interaction with a residue present in CB2 (Ser165) that is absent in CB1 and therefore could lead to selectivity for CB2. This was performed using the 'docking' protocol within Spark™.5 Upon investigation of the binding modes, it was observed that the cyano group in ligand AM841 extended into a spacious cavity within the CB2 binding pocket (Figure 2).
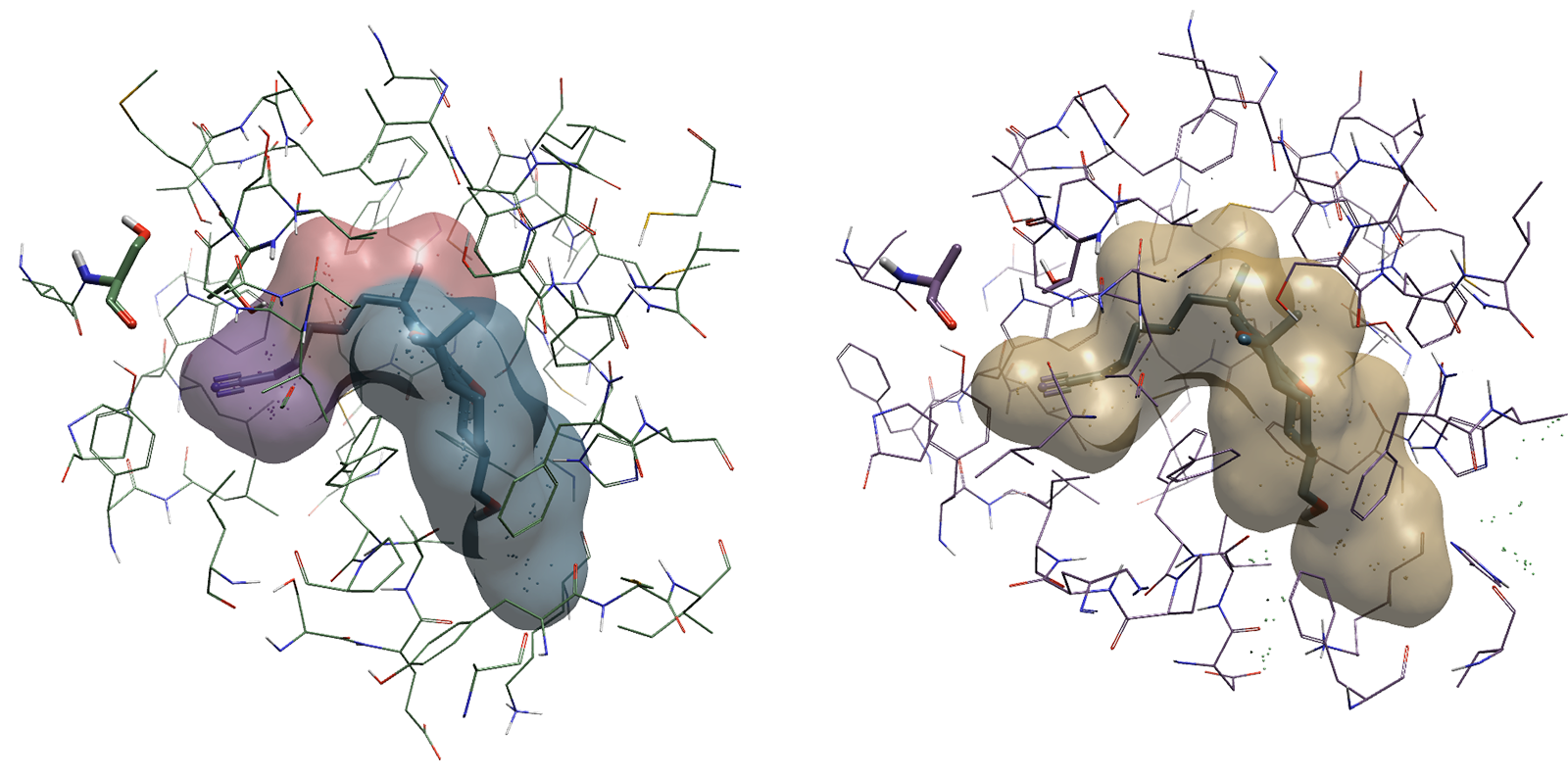 Figure 2. Binding pockets of CB2 (left) and CB1 (right) showing the Ser165 residue present near the binding cavity in CB2 as opposed to Ala248 in CB1.
Figure 2. Binding pockets of CB2 (left) and CB1 (right) showing the Ser165 residue present near the binding cavity in CB2 as opposed to Ala248 in CB1.
Ligand AM12033 was taken forward to perform an R-group replacement experiment using the binding pocket of CB2 as an excluded volume and searching for fragments from ChEMBL and SureChEMBL (very common and common fragments).
Analysis of the Spark results require triaging to remove compounds which are unsuitable for the project under investigation. Careful consideration of the parameters utilized in the triaging step is needed through application of medicinal chemistry and biological understanding of the target. Firstly, in order to prevent adversely affecting entropic penalties on binding, flexibility was restricted to a maximum of 5 rotatable bonds. The number of rings was considered as a way to mitigate this but had to be carefully balanced to prevent making the molecule too hydrophobic, therefore a cut off of no more than 4 rings was applied. Finally, ligands that contained groups that are chemically intractable, were filtered out. Docking was performed on the remaining ligands, to generate binding poses of the selected ligands in both the CB1 and CB2 receptors. At this stage, additional checks could be performed such as removing ligands that contain problematic groups (for example, PAINS).
Once the binding poses of each ligand were obtained for both receptors, a Python script utilizing the Python Flare API (pyFlare) was used to triage the results of the ligand docking experiment. The script calculates the Electrostatic ComplementarityTM (EC)6 and Lead Finder (LF VSscore)7 score for each ligand in each of the two receptors and calculates the difference between the two values (where diff = CB1 score – CB2 score). A more negative value represents selectivity towards CB2 as opposed to CB1 enabling filtering of the ligands to focus on those with favorability towards CB2. Based on the data range for each score across all ligands, the results were filtered with a cut-off value of 3 for LF score difference and 0.3 for EC score difference applied (meaning only ligands with a difference in LF score of 3 or more and EC 0.3 or more between CB2 and CB1 were kept).
After triaging the results, Cresset’s Discovery scientists used their knowledge of CNS active ligands to filter out unlikely candidates. Ligands that contained: anions, ligands with over 3 hydrogen bond donors, over 7 hydrogen bond acceptors, over 8 rotatable bonds, and highly hydrophobic ligands (ligands with a predicted LogP of 3 and above) were removed.8 At the end of this step, a single representative compound that satisfied all of the desired conditions was chosen as the new reference starting point (compound 1) for virtual screening utilizing Blaze™.9
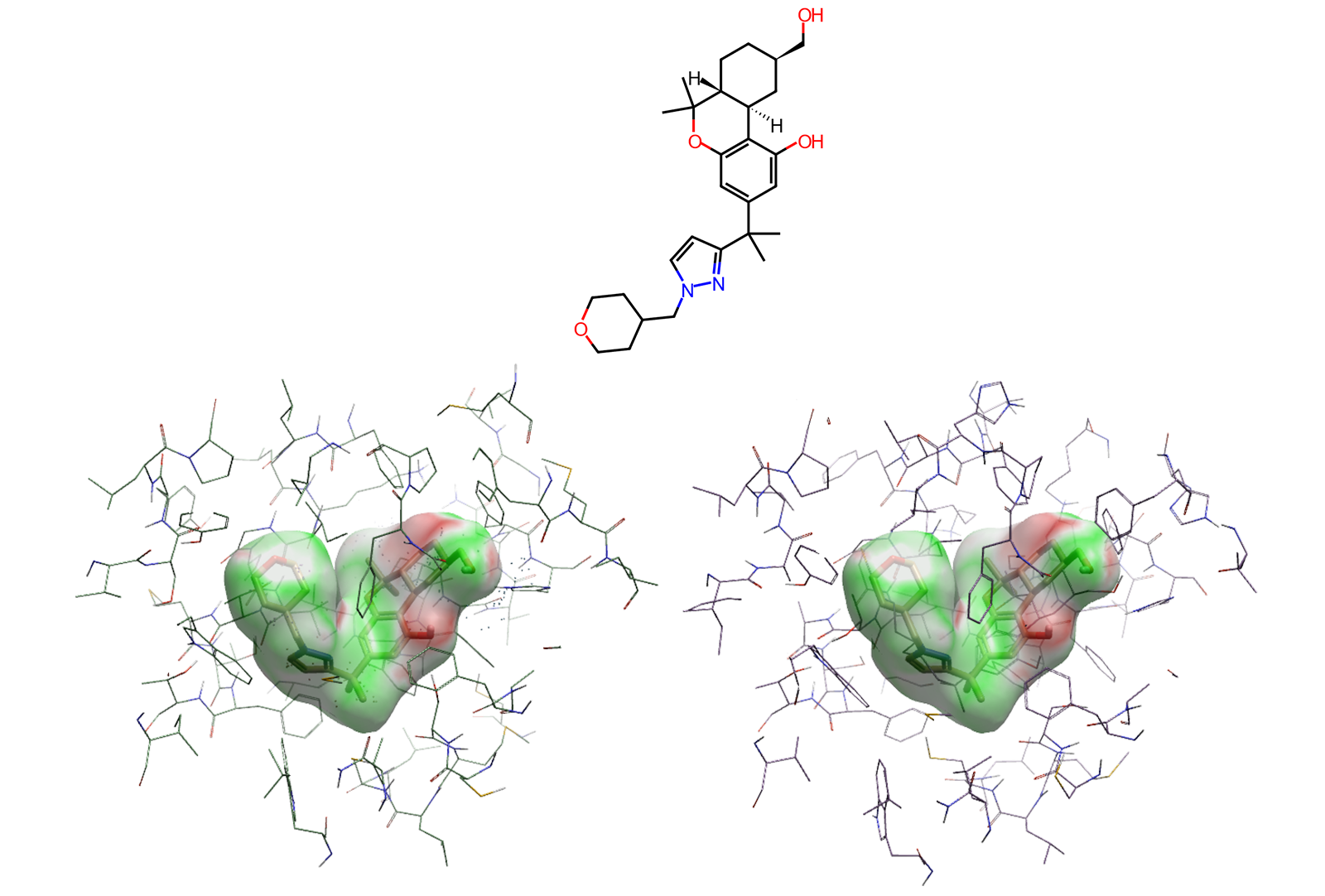 Figure 3. Compound 1 in 2D (above) and 3D with EC to protein surface visible in both CB2 (bottom left) and CB1 (bottom right).
Figure 3. Compound 1 in 2D (above) and 3D with EC to protein surface visible in both CB2 (bottom left) and CB1 (bottom right).
Both this reference molecule (compound 1, Figure 3) and AM12033 were used as reference templates in independent virtual screens, searching the ChEMBL, Enamine Advanced, Enamine HTS, Enamine Premium, Molport and WuXi Screening databases. CB2 was used as excluded volume for these screens.
The results were then filtered and anions removed. The top 10,000 hits were triaged using the previously defined criteria, utilizing the custom pyFlare script. The remaining hits were redocked into CB1 and CB2 and the differences in EC and LF score calculated. These results were plotted in histograms to visualize the metrics from the virtual screening of both compound 1 and AM12033.
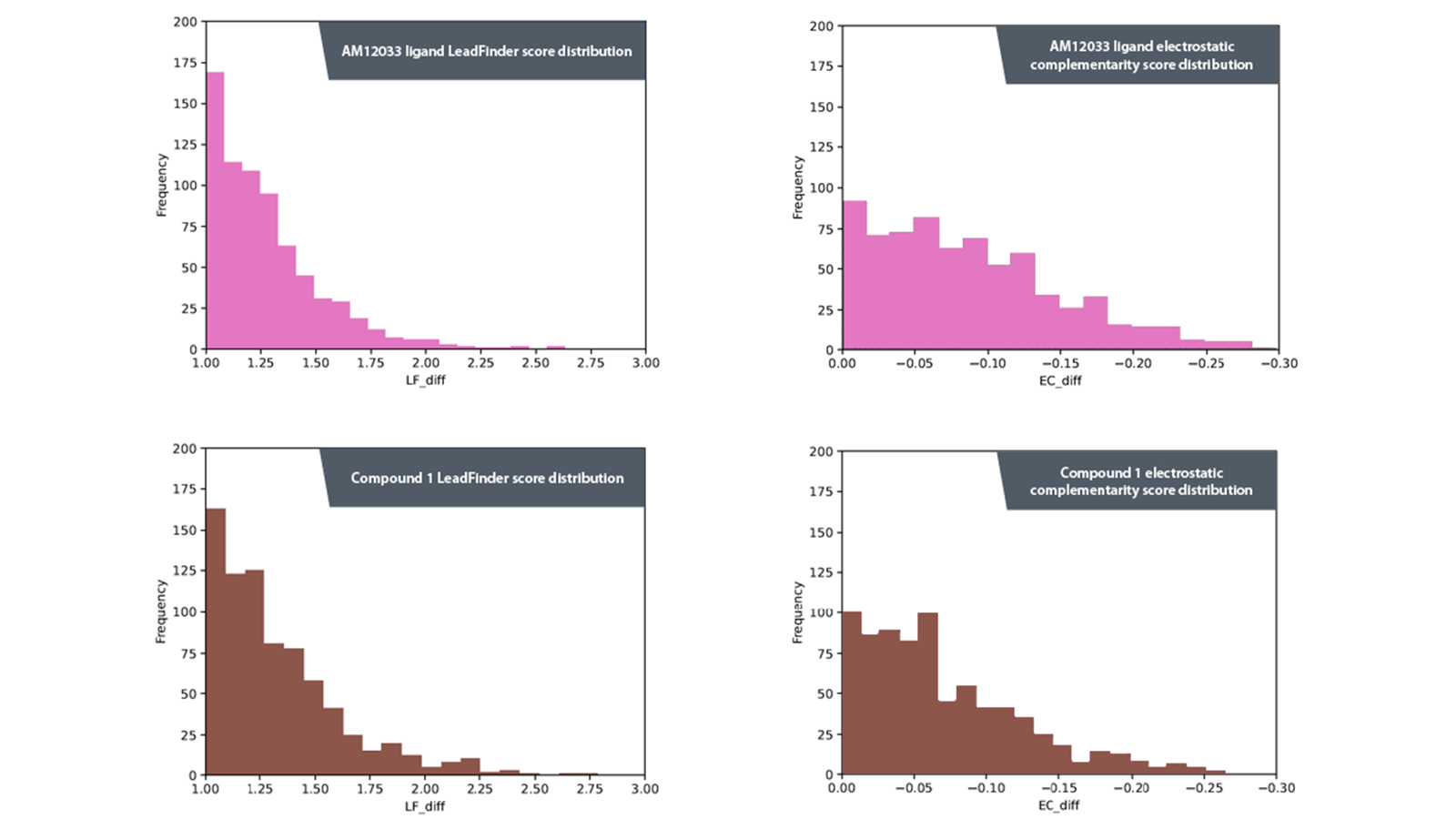
Figure 4. Histograms showing distribution of differences between LeadFinder (LF) and Electrostatic Eomplementarity (EC) scores in virtual screening results between receptors. Top left AM12033 ligand LF score distribution. Top right AM12033 EC score distribution. Bottom left compound 1 LF score distribution. Bottom right compound 1 ligand EC score distribution.
As seen in Figure 4, compound 1 produces further right-skewed histograms than AM12033, demonstrating that prework before a virtual screen can be beneficial for finding suitable starting points for receptor subtype selective ligands.
Analyzing the graphs shows that using compound 1 produced results where a higher proportion of ligands had a selectivity bias towards CB2 than CB1 for both the docking scores and the EC scores. A difference in docking score of less than 1 or in the negative range indicates selectivity toward CB1 and, since the aim of this study was to identify compounds that were selective to CB2, the results in this negative range for compound 1 were discounted. The remaining compounds then underwent a thorough visual analysis using medicinal chemistry knowledge to identify a final list of compounds that could be purchased or synthesized for biological testing.
Figure 5 shows four examples of high-ranking molecules following the final triaging step. The experiment has provided a collection of diverse molecules that all display selectivity towards CB2 as well as satisfy the constraints applied using knowledge of the target, its location and synthetic chemistry.
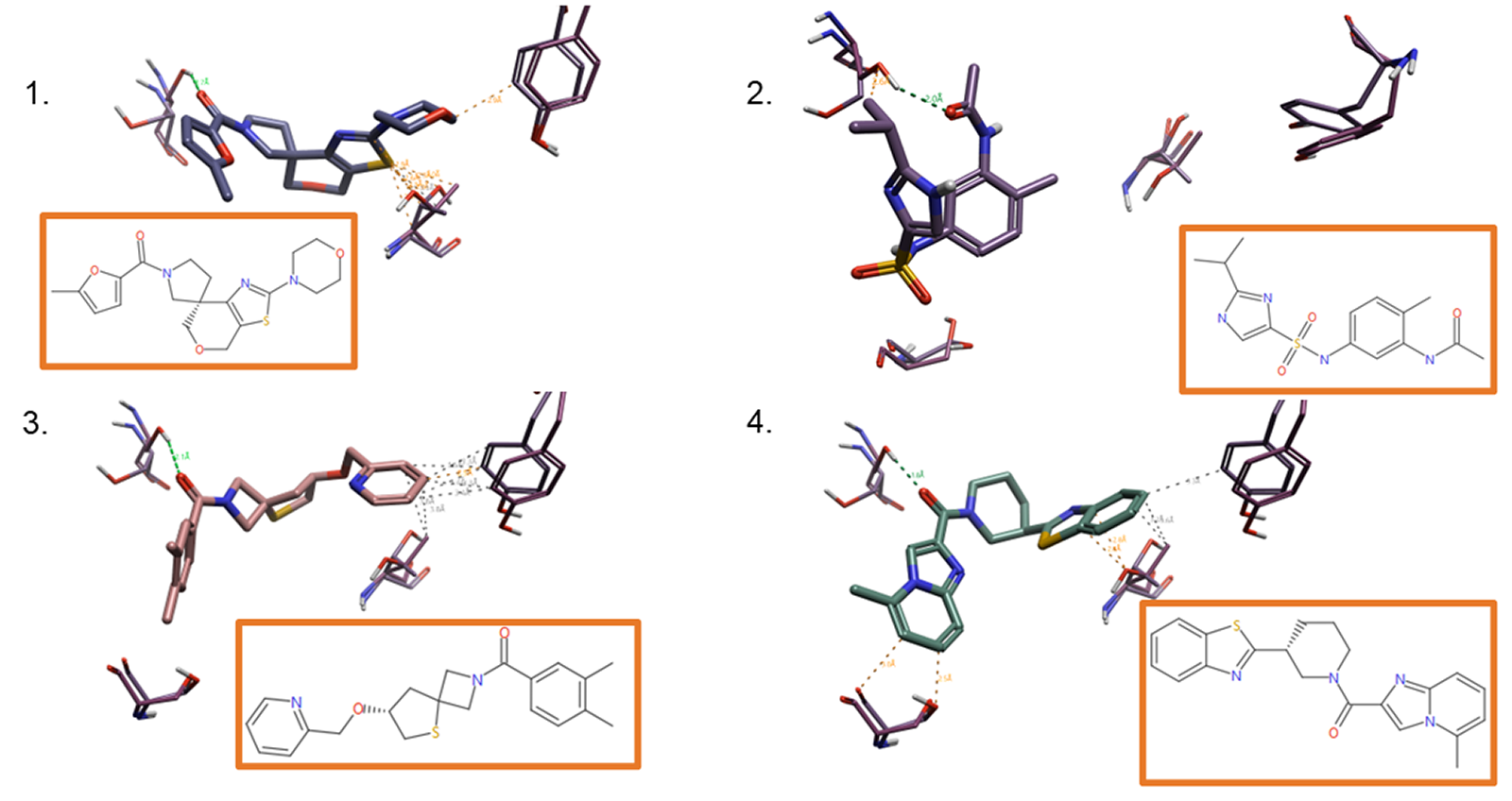
Figure 5. Examples of the top-ranking ligands following the triaging steps, in 2D and 3D.
Learn more in our webinar
For a visual demonstration of the steps performed in this workflow, watch our webinar 'Identifying leads with targeted selectivity through virtual screening - optimizing the starting point to predict the molecules most likely to succeed' – request webinar recording.
Contact Cresset to accelerate your project using our CADD Software or Discovery CRO
Cresset provides software and discovery services to supercharge your research efforts. Our expert team has a breadth of experience across medicinal and computational chemistry, and biology to work alongside you to deliver your project goals, while our software provides cutting-edge methodology to provide valuable insights into your project.
To learn more about how we can support you with a virtual screening project, request a CADD software evaluation or request a confidential discussion with our Discovery CRO team.
References
- P. F. U. G. A. M. M. D. G. E. Rossi F, "Role of Cannabinoids in Obesity," Int J Mol Sci., 2018
- S. A. M. A. H. D. O. B. Verty AN, "Anti-Obesity Effect of the CB2 Receptor Agonist JWH-015 in Diet-Induced Obese Mice," PLoS One, 2015
- Flare™, version 7.1, Cresset®, Litlington, Cambridgeshire, UK; https://www.cresset-group.com/flare/
- https://www.cresset-group.com/discovery/specific/protein-preparation/
- T. e. a. Hua, "Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-Gi Complex Structures," Cell, 2020, vol. 180
- Spark™, version 10.7, Cresset®, Litlington, Cambridgeshire, UK; https://www.cresset-group.com/spark/
- Flare™, Cresset®, Litlington, Cambridgeshire, UK; https://www.cresset-group.com/flare/; Bauer M., Mackey M.; Electrostatic Complementarity as a Fast and Effective Tool to Optimize Binding and Selectivity of Protein–Ligand Complexes J. Med. Chem. 2019, 62, 6, 3036-3050
- Lead Finder™, version 7.1, BioMolTech®, Toronto, Ontario, Canada; https://www.cresset-group.com/lead-finder/
- H. L. G. Pajouhesh, "Medicinal chemical properties of successful central nervous system drugs," Neurotherapeutics, 2005, 541–553
- Blaze™, version 10.3.2, Cresset®, Litlington, Cambridgeshire, UK; https://www.cresset-group.com/blaze/