Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Last year we announced our intention to develop a new structure based design application based on key research projects that we have been working on at Cresset. We have showed that our XED force field has significant advantages in the calculation of electrostatic protein interaction potentials and the calculations of water positions and energetics using RISM.
To bring this new science to you presented us with a significant challenge. Our existing ligand-based design products work well for ligands but lack the necessary data structures to support structure based approaches and would have required significant re-design to retro-fit them. Accordingly, we chose to build a new application that will not only provide the framework for our new science but will also be a receptacle for future innovations, both our own and those of leading academic groups.
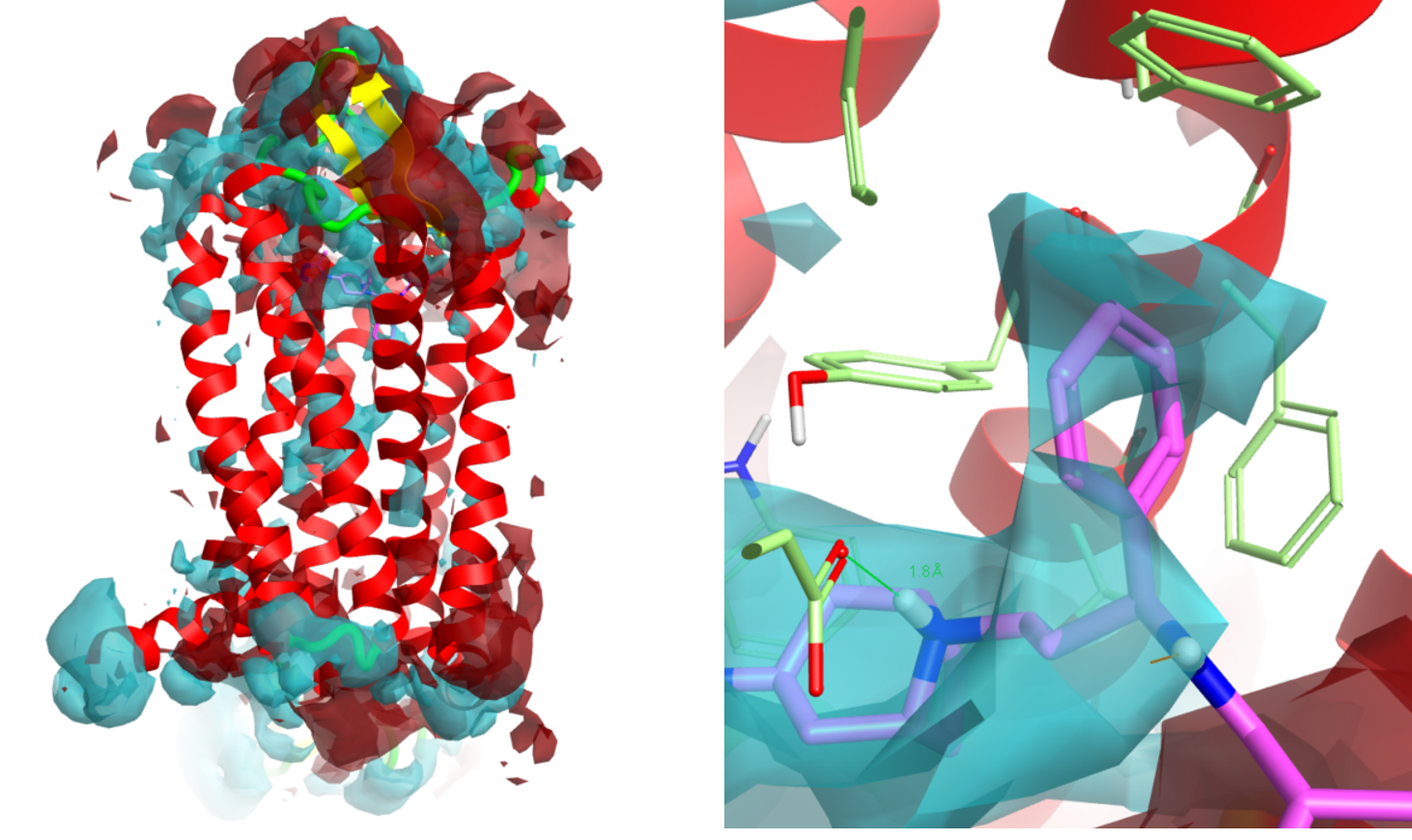
Figure 1. Protein interaction potentials (red=positive, blue=negative) for PDB 4MBS; (a) for the entire structure highlighting the helix dipoles and (b) in the ligand binding region showing the negative potential derived from GLU283 and the aromatic box encasing the ligand’s pendent phenyl group.
The last six months have seen significant progress towards our goal of a fully functional, released product by spring 2017. We now have in-house an application that includes primitive interfaces to most of the cutting edge scientific approaches that we had identified for the release version as well as to the Lead Finder docking engine. This alpha version is undergoing extensive validation and is sufficiently functional to be of active use by Cresset Discovery Services.
However, our long term aim is to provide exciting new science while retaining the simplicity of user interface and ease of use. Significant work remains: we still have to incorporate the last few algorithms and extend the interfaces to the new science ready for a full beta test release in early in 2017. If you are interested in taking part in the beta release early in 2017, then please contact your account manager.
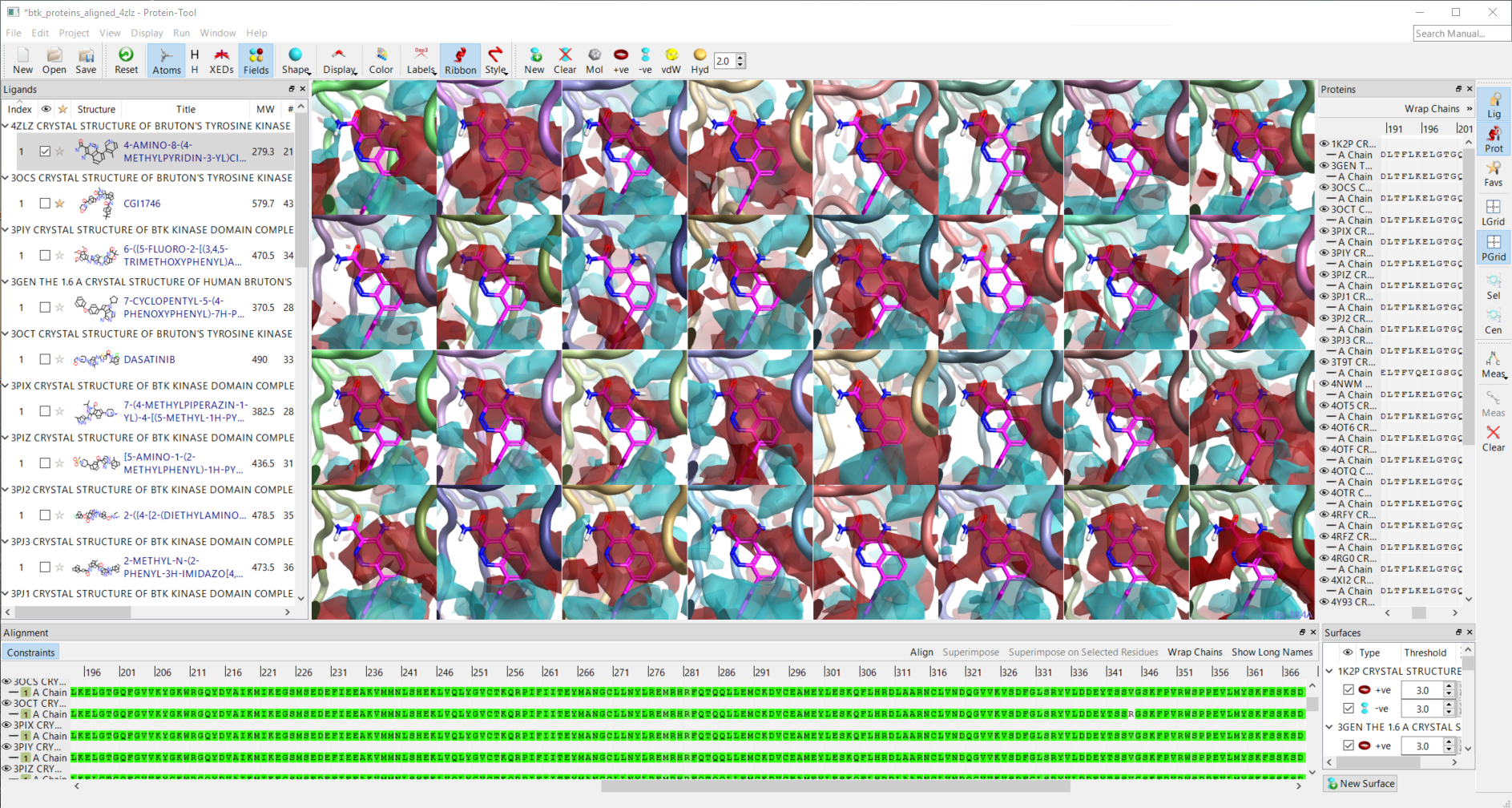
Figure 2. Screenshot of the new application showing protein interaction potentials for 32 BTK proteins in the RCSB. Proteins were aligned within the application using COBALT and superposed by matching C-alpha atoms. The ligand of pdb 4zlz is shown for reference. A positive interaction potential (red) is present in most cases indicating the preference for an electron rich ligand.
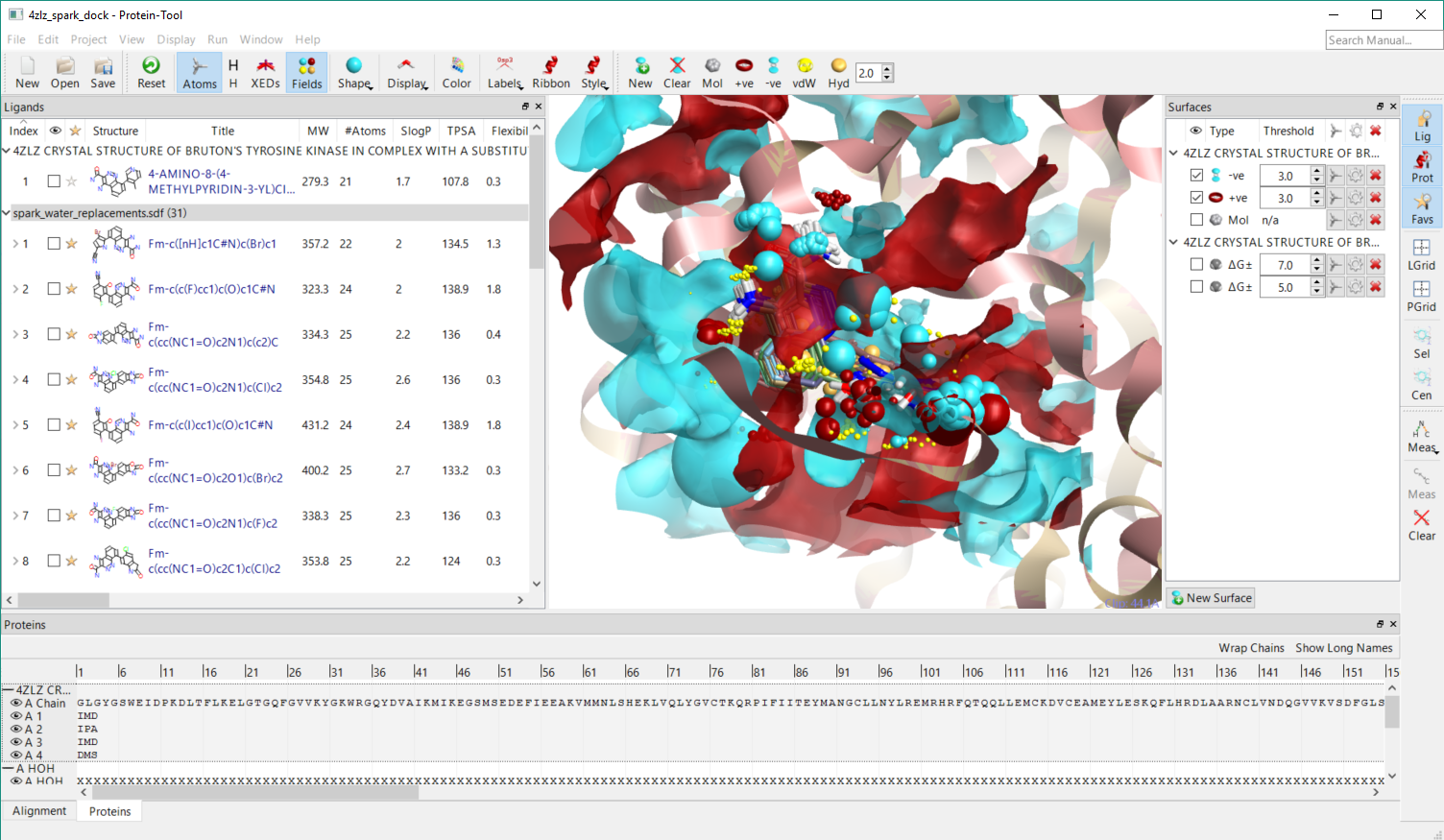
Figure 3. Spark results for replacement of a pyridine-water complex docked to pdb 4zlz using Lead Finder within the structure-based design application and shown with the protein interaction potential and ligand field points.