What are PROTAC molecules and how do they work?
PROTAC®s (PROteolysis TArgeting Chimeras) are heterobifunctional molecules typically consisting of three components: a ligand targeting the Protein Of Interest (POI) for proteasomal degradation, a ligand targeting an E3 ligase protein, and a linker to connect them. By utilizing this bifunctional approach, the POI is marked for degradation by the body’s native degradation pathways (Figure 1). This complex system can enable the treatment of biological targets previously considered 'undruggable' by small molecule therapeutics.1 These heterobifunctional molecules have many advantages over traditional small molecule inhibitors, such as their inherent sustained binding, which may allow for less frequent dosing, and their activity in inhibitor-resistant cancer models due to their bifunctional mode of action. With their numerous advantages, PROTACs are a popular and promising avenue of research.2 However, due to their size and complexity, the design of PROTACs with suitable drug-like profiles poses new challenges.
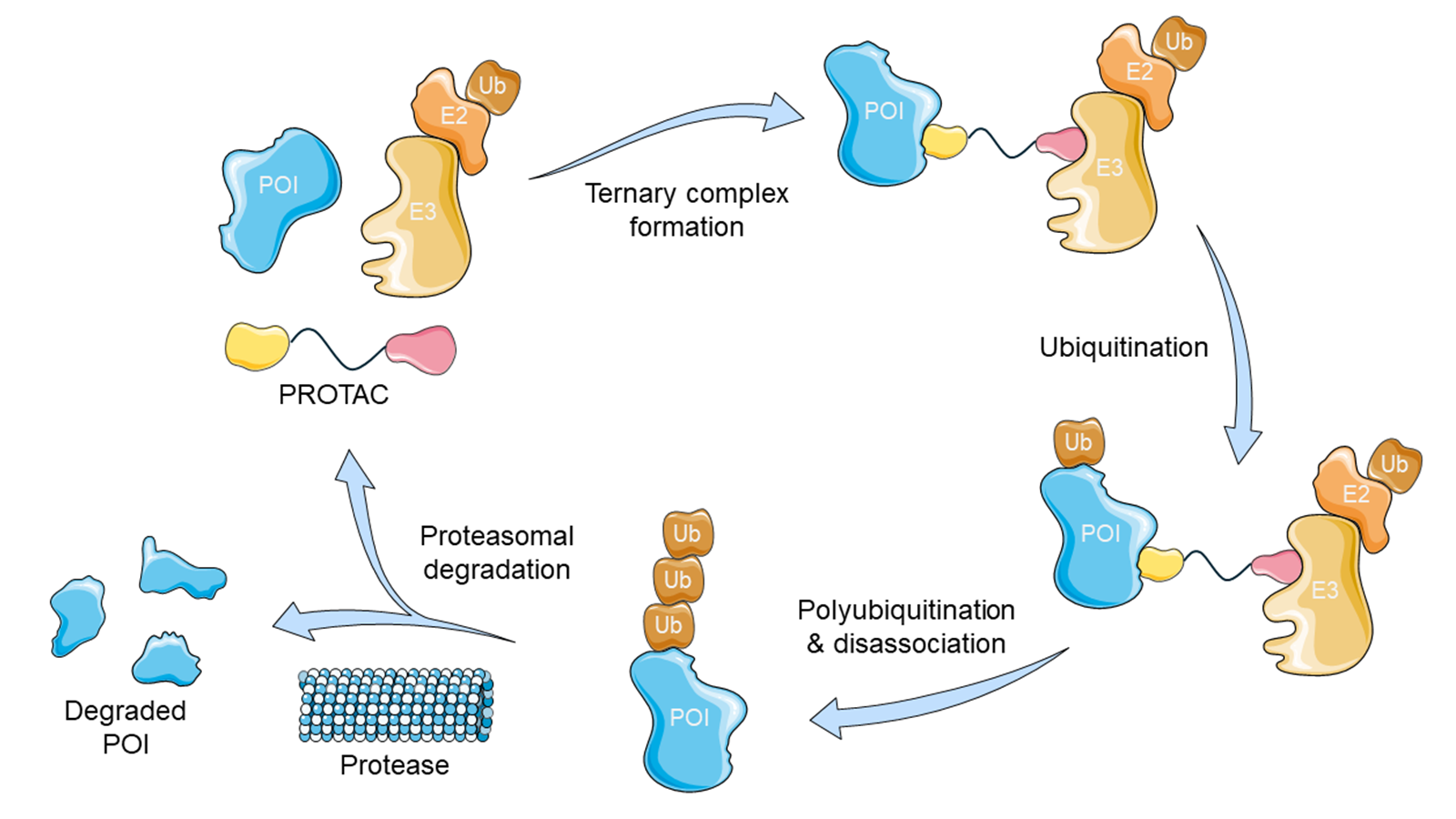
Figure 1. The PROTAC degradation cycle. (Top Left) The POI binder of the PROTAC binds to the POI, whilst the E3 binder of the PROTAC binds to the E3 ligase. (Top Right) The POI-PROTAC-E3-E2-Ub ternary complex forms. (Bottom Right) The POI is polyubiquitinated by the intermediate complex, marking it for degradation by the proteosome. (Bottom Left) The POI is degraded and the PROTAC is released, allowing it to act again. This figure was partly generated using Servier Medical Art, provided by Servier, licensed under a CC BY 4.0 license (CC BY 4.0 Deed | Attribution 4.0 International | Creative Commons).
Using molecular modeling to inform the design of new PROTAC molecules
Computational methods play an important role in the drug design process. Through modeling approaches which enable us to visualize and design compounds with complementary interactions to their intended targets, the drug design pipeline can be made more efficient. Some of these approaches can be applied to PROTAC design to effectively tackle the challenges that these bifunctional molecules present. In this article, we will be looking at one of the ways in which CADD workbench Flare™ can aid PROTAC design.
Cresset Field Points3 can be a useful guide to enhance compound design. These points are the local extrema of electrostatic, van der Waals and hydrophobic fields calculated with Cresset XED force field (further information can be found on the Cresset Science Overview webpage). By using these as descriptors of bioactivity, molecules can be modified in ways that optimize their interaction with a chosen target. As such, Field Points can show how changes to molecular structure, such as changes to a PROTAC linker, will affect the interactions of compounds to their biological target.
As an example, we will be looking at a PROTAC targeting STAT5 (Signal Transducer and Activator of Transcription 5), a signaling hallmark of acute myeloid leukemia. This target has been historically difficult to drug with small molecule inhibitors, and those with desirable potency and selectivity for STAT5 have hitherto not been found. One particularly potent and selective PROTAC degrader of STAT5 is AK-2292 (Figure 2).2
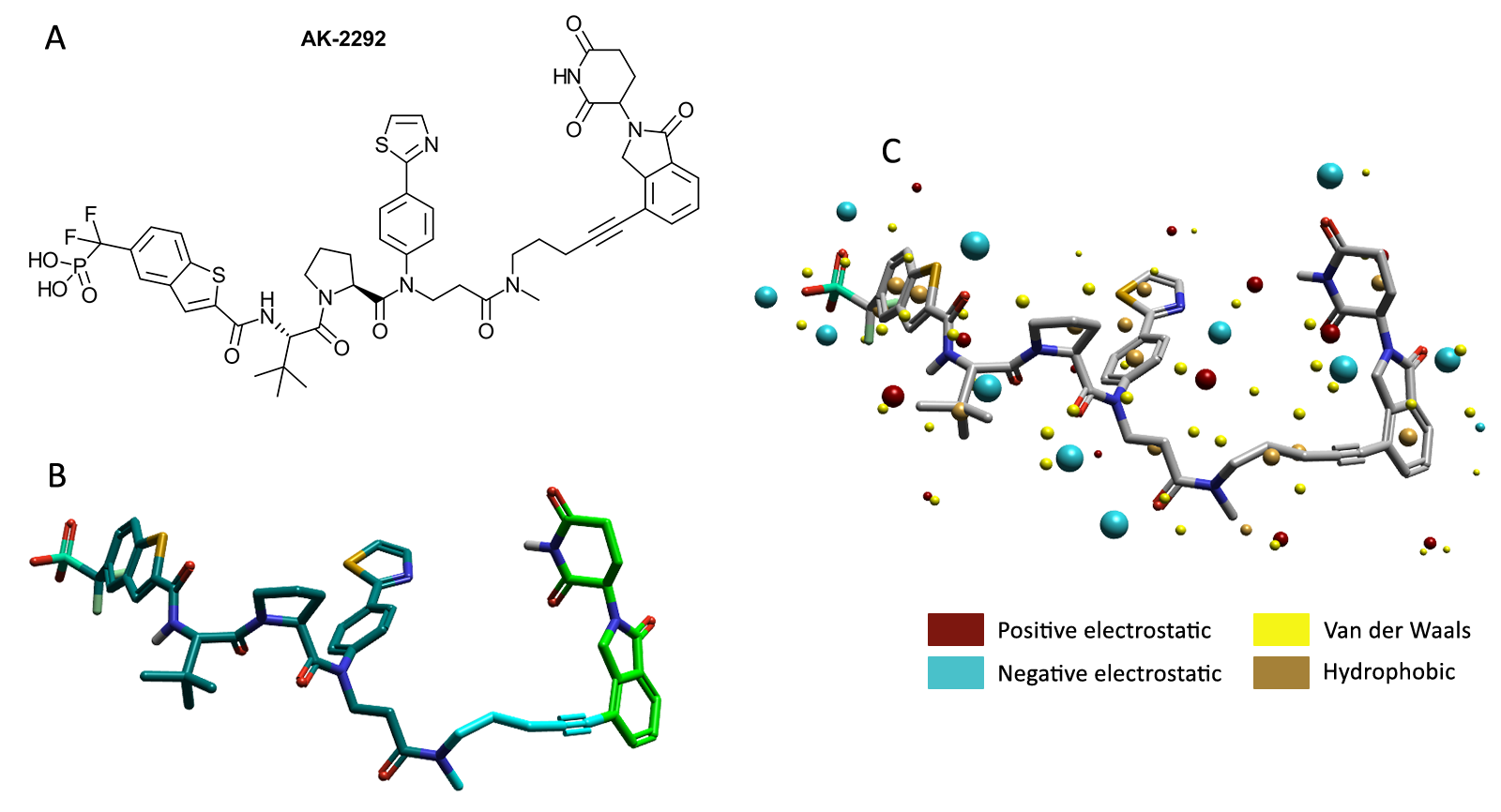
Figure 2. (A) 2D structure of AK2292. (B) Bioactive conformation of AK2292 taken from a crystal structure (PDB: 7TVA). The STAT5 binder (dark green), linker (cyan) and CRBN binder (light green) can be seen. (C) Cresset Field Points have been generated around the molecule and displayed using Flare.3
AK-2292 consists of a POI binding ligand, created from a small molecule inhibitor of STAT5, a CRBN binding ligand, and a linker connecting the two ligands. In their study, the authors tested various linkers and measured the change in degradation activity, culminating in the creation of AK-2292, which showed potent and selective degradation of STAT5.2
Optimizing PROTACs using Hit Expander
In this experiment, we will use Flare’s Hit Expander tool to further explore linker modifications. Hit Expander allows simple functional groups to be added to amenable positions on a ligand or on a chosen portion of it, as well as perform aromatic single atom substitutions (Figure 3).
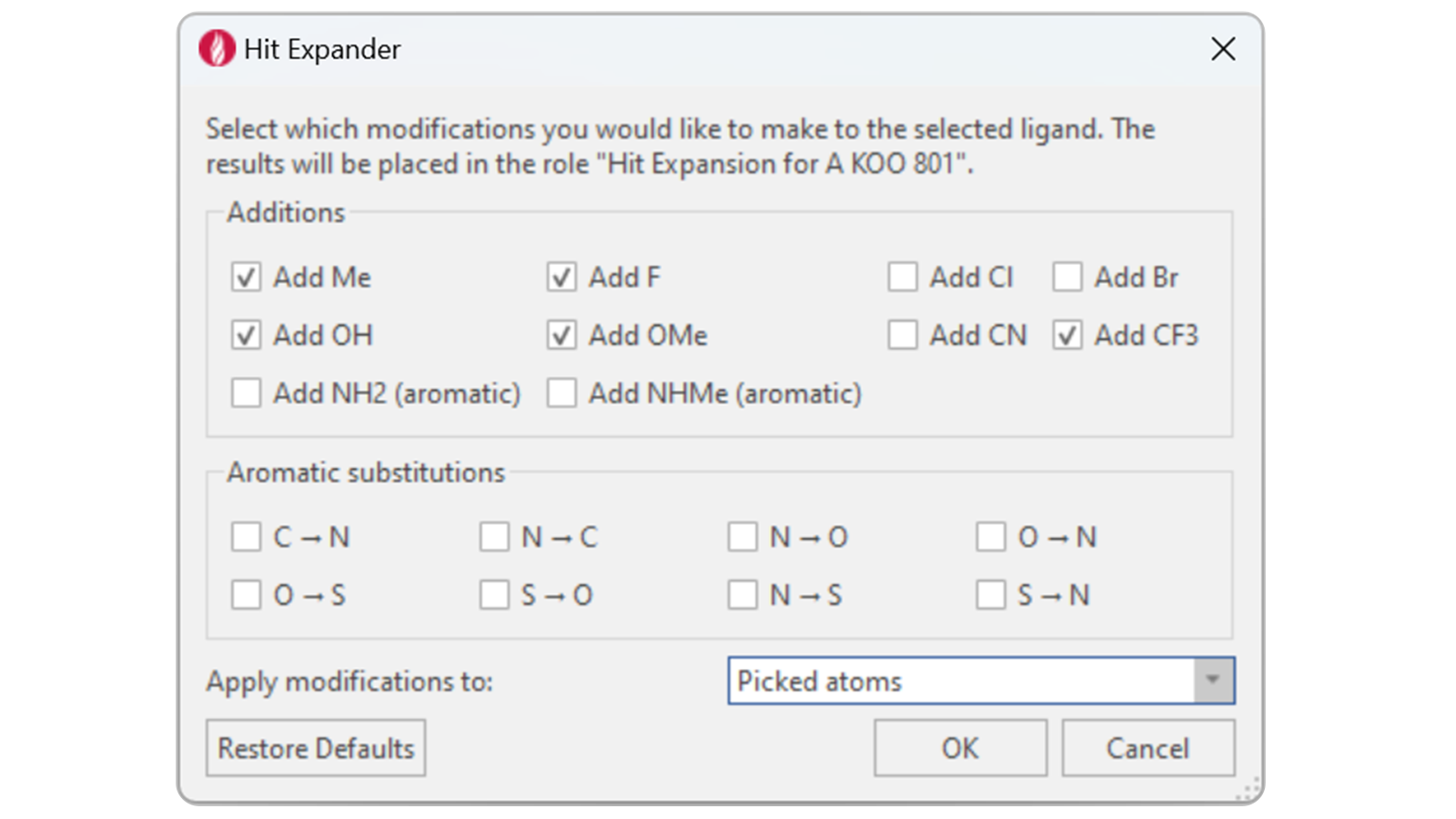
Figure 3. The Hit Expander window showing the parameters chosen for this experiment.
To obtain modifications, the linker portion of the AK-2292 PROTAC was selected in the Flare 3D view, and several groups were chosen as additions in the Hit Expander window: methyl, fluoro, hydroxyl, methoxy, and trifluoromethyl groups: the output yields 35 new variants of AK-2292.
Although only small substitutions were introduced with Hit Expander, these might have a significant impact on the molecular fields and field points of the linker. In order to further refine and optimize the 3D structure and electrostatic description of the substituted ligands, the 35 variants were re-aligned to AK-2292 using ligand-based substructure alignment in Flare.
Two particularly interesting results are shown in Figure 4 with field points and with full ligand fields, both calculated in Flare using the XED force field.
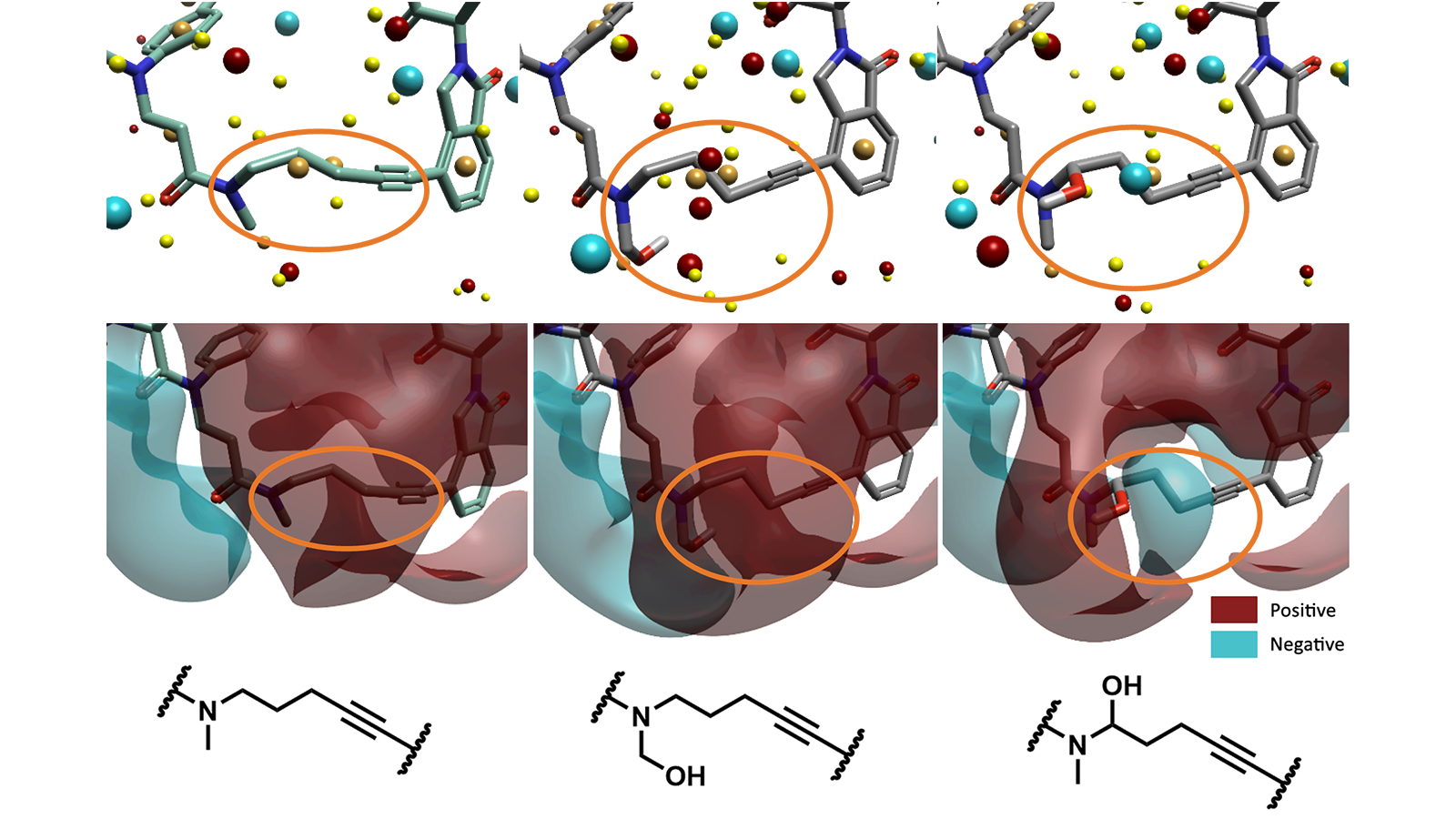
Figure 4. The crystal structure ligand (left, AK-2292) and two interesting Hit Expander results. The linker portion of the molecules is highlighted with an orange circle. Along the top Cresset Field Points are displayed and along the middle ligand fields are displayed.
We can see that the ligand fields surrounding the linker portion of our PROTAC molecules have changed in the chosen Hit Expander results (Figure 4). This may be beneficial in many ways, such as improving binding to the target protein or solubility, a known issue with large, greasy linkers that are typical of PROTACs. Hit Expander can provide synthetic ideas for alternative linkers, and therefore used to improve PROTACs druggability.
The new molecules generated by Hit Expander can now be used to perform additional experiments, such as docking or analysis of the Electrostatic Complementarity™ with the target protein.4 With the additional compound profiling the molecules can be further optimized, allowing rational design and improved performance of their PROTACs.
Hit Expander can generate new ideas for your PROTAC linkers
Hit Expander is a powerful yet easy-to-use tool for PROTAC design. Linker adjustment is often a challenging part of developing potent and selective PROTAC molecules. The Hit Expander tool allows the user to explore new ideas for improving compound properties or protein interactions with just a few clicks, aiding in the development of highly active and selective complexes.
References
- Guedeney, N., Cornu, M., Schwalen, F., Kieffer, C. & Voisin-Chiret, A. S. PROTAC technology: A new drug design for chemical biology with many challenges in drug discovery. Drug Discovery Today 28, 103395 (2023).
- Kaneshige et al. Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia. Journal of Medicinal Chemistry 66, 2717–2743 (2023).
- Cheeseright, T., Mackey, M., Rose, S. & Vinter, A. Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation. J. Chem. Inf. Model. 46, 665–676 (2006).
- Electrostatic ComplementarityTM scores: How can I use them? https://www.cresset-group.com/about/news/ec-scores-how-use/



