Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Macrocyclization of pharmaceutical compounds plays an increasing role in drug discovery. Macrocycles can provide several advantages such as favorable drug-like properties, and increased selectivity and binding affinity. Here we present a case study of designing macrocyclization strategies for reported BRD4 inhibitors with Spark™1, Cresset’s bioisostere replacement and scaffold hopping tool.
Macrocyclization of pharmaceutical compounds can provide several advantages, such as diverse functionality, favorable drug-like properties, and increased selectivity.2 Also improved binding can result from macrocyclization by locking the molecule in a low-energy binding conformation and reducing entropic cost upon binding. 2–4 One challenge in harnessing the advantages of macrocyclic compounds is the difficulty in synthesizing such molecules. Prediction of effective macrocyclization strategies prior to synthesis is, therefore, crucial for successful macrocycle drug discovery.
Recently, Wang et al. reported a new class of pyridone-based BRD4 bromodomain inhibitors that show promising anticancer effects in cell lines and xenograft models.4 Starting from a pyridone fragment hit, the compound was further optimized using structure-based design to achieve one-digit nanomolar potency. Macrocyclization further improved binding affinity, cellular efficacy, and pharmacokinetic compound properties (Figure 1). 4
In this case study, we used Spark, Cresset’s bioisostere replacement and scaffold hopping tool, to design macrocyclization strategies for non-macrocyclic, pyridone BRD4 inhibitors and evaluate results against experimental data reported by Wang et al.
Figure 1: Development of pyridine-based BRD4 bromodomain inhibitors reported by Wang et al. Macrocyclization of optimized fragment [2] improved binding affinity (Ki values from TR-FRET binding assay) about 4x. In addition to improved binding, compound [3] shows an increased cellular efficacy and improved pharmacokinetic properties.
Spark replaces a specified moiety in a given starter molecule with fragments that exhibit similar electrostatic and steric properties.5 Spark’s molecular comparisons are based on molecular fields, computed by the XED force field.6–8 A major benefit of Spark is that it retains a larger amount of 3D information in the search for replacement fragments: instead of scoring only the respective replacement fragment in isolation, the whole final molecule is scored, considering any electronic and conformational effects the new scaffold may have on the rest of the molecule. Replacement fragments are selected from Spark’s fragment databases which are derived from commercially available and literature compound collections. After identifying fragments that contain the required number of attachment points and possess a shape / geometry compatible to the starting molecule, the molecules are minimized using the XED force field to remove bad clashes and unfavorable geometries. A final molecular field calculation and similarity optimization is then performed to yield a final score.
The macrocyclization wizard was used to design BRD4 macrocycle inhibitors. The wizard module provides a quick and easy-to-use GUI workflow with dedicated Spark settings tailored for macrocycle design (Figure 2). For this experiment, we used the X-ray structure of BRD4 in complex with the non-macrocyclic compound [2] (pdb 5UEY) as receptor, and compound [2] as the starter molecule (Figure 2). The bioactive conformation of [2] shows that the ethoxyl group of the pyridone scaffold is in close proximity to the 2,4-difluorophenoxy ring (Figure 3). Additionally, the ligand is partially solvent exposed in this region of the binding site, offering sufficient space to accommodate additional linker atoms. Therefore, we chose to link point 1 (OEt moiety) with point 2 (hydrogen atom) at position 6 of the 2,4 difluorophenoxy ring (Figure 3).
Figure 2: Selection of attachment points in the macrocycle wizard. Attachment point 1 is the ethoxy group and attachment point 2 is hydrogen 6 on 2,4 difluorophenoxy ring. The 3D window (right side) visualizes attachment points and confirms correct selection.
Figure 3: Inhibitor [2] in complex with BRD4 (pdb 5UEY). The close proximity between the ethoxy group and the 2,4 difluorophenoxy ring and the partially solvent exposed volume in this area suggests that linking these features may be feasible.
To bias linker design towards existing chemistry, oxygen was specified as attachment atom for attachment point 1. The second attachment point was left unconstrained. Additionally, the receptor structure was used as an excluded volume to guide linker design and the field points of carbonyl and sulfonyl groups forming hydrogen bonds with the receptor were constrained. The experiment was run with the dedicated ‘Ligand Joining / Macrocyclization’ settings against the fragment databases CHEMBL_common, Common, and VeryCommon, containing altogether about 120K fragments. Results were evaluated in Spark and molecular overlays were rendered in Flare™9.
The macrocycle wizard experiment generated 500 rank-ordered macrocycles derived from fragments merged with the starter molecule. The X-ray structure of the reported and structurally validated macrocycle [3] (Figure 4) shows that upon cyclization the molecule retains its key interaction with the BRD4 pocket, however, compound [3] adapts a slightly different conformation than [2], especially for the 2,4-difluorophenoxy ring. Despite this conformational change, the top scoring Spark results contained several macrocycles closely related to [3] (Figure 5), such as compounds with a butanol (rank 11), propan-1,3-diol (rank 9), 4-aminobutan-1-ol (rank2), butan-2-ol (rank 4), or a 3-buten-1-ol linker (rank 17). Compound [3] was found at rank 59 (Figure 5).
Among the top Spark results, linkers with 4 to 6 atoms were particularly enriched (Table 1). This finding is in good agreement with the experimental data reported by Wang et al., who found that short macrocycle linkers with only 3 atoms decrease the binding affinity by about 50x compared to a 5 atom linker.
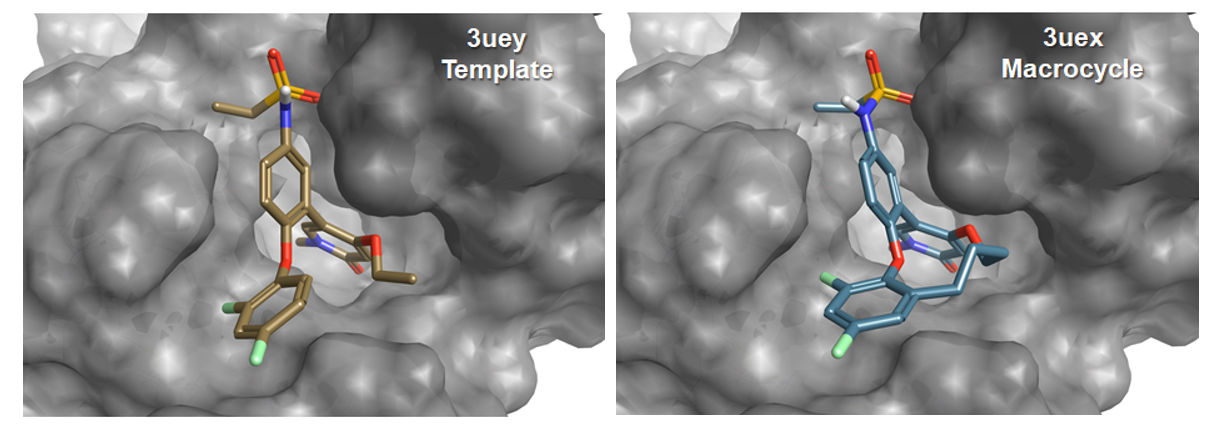
Figure 4: X-ray structures of BRD4 in complex with [2] (3UEY) and the experimentally validated macrocycle [3] (3UEX).
Figure 5: Spark macrocyclization results. Among the top scoring results, Spark designed several linkers that are either identical or very similar to compound [3]. ECFP4 values are Tanimoto similarities to [3]. Scores were calculated by Spark and used for ranking of results. Attachment points A1 and A2 as specified in the method section.
Four atom linkers were only slightly less potent (factor 3) than the 5 atom linker, whereas 6 atom linkers showed a comparable binding affinity.4
The top ranked Spark results for each linker size are shown in Figure 6. Interestingly, for the shorter linker sizes (e.g., butan-2-ol with 4 linker atoms) Spark predicted fragments that mimic the hydrophobic volume of the ethoxy side chain of [2]. Omission of parts of this hydrophobic volume may be contributing factors in the observed potency loss for 3 atom linker macrocycles. Adding a Me group to the linker to address this feature of [2] (e.g., compound at rank 6 in Figure 6) may, therefore, result in short macrocycles with improved potency against BRD4.
Table 1: Distribution of linker sizes in top10, top25, and top50 results from Spark (500 total).
| Top N results | 3 linker atoms | 4 linker atoms | 5 linker atoms | 6 linker atoms | 7 linker atoms |
| 10 | 2 | 6 | 1 | 1 | 0 |
| 25 | 2 | 16 | 4 | 8 | 0 |
| 50 | 2 | 32 | 12 | 9 | 0 |
Figure 6: Spark macrocyclization results. Among the top 10 results Spark designed compounds with linker sizes between 3 to 6 atoms. The top ranking result for each linker size is shown. Attachment points A1 and A2 as specified in the method section.
This case study demonstrates that Spark can successfully design macrocycles that are identical or very similar to reported BRD4 macrocycle inhibitors.4 The distribution of generated linker sizes was in good agreement with experimental SAR data.4
The Spark macrocyclization wizard is a quick and easy-to-use workflow that generates meaningful and diverse design ideas that can guide macrocycle drug discovery.