Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Flare 2.0 comes with a full-featured Python API which enables both high-level access to scientific functionality and low-level access to the graphical user interface and internal processes. In addition to Flare methods, you will have access to RDKit, NumPy, SciPy, and to virtually any other Python module that can be pip-installed. RDKit molecules are treated just as native Flare molecules, and can be loaded interchangeably to Flare’s ligand table. This means that you can generate a virtual 2D library using RDKit reaction SMARTS and then turn it into a 3D library of field-enabled Cresset molecules without ever leaving the Flare environment.
Additionally, if you realize that you often use a certain Python script as part of your Flare workflow, you may decide to associate it to a custom control in the Flare ribbon user interface, be it a button, a menu, or a complex dialog with signals and callback functions (Figure 1).
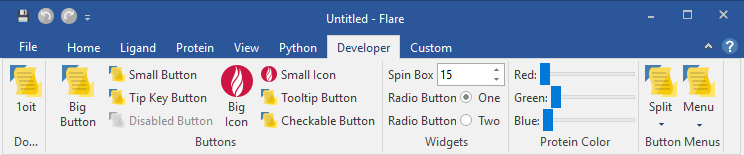
We have done our best to write a clear and easy-to-browse HTML documentation, but even more importantly we have come up with a number of sample scripts which cover a variety of use cases, in order to get you started as quickly as possible.
Python commands can be loaded and executed in Flare in a number of different ways, ranging from highest automation to highest interactivity.
For example, when you need to carry out a completely automated task, such as overnight preparation of a panel of proteins followed by docking of several ligand series, the most convenient option is to write a Python script that runs outside the Flare GUI, such that it can be distributed on a cluster via a queueing system for maximum performance. This use case is covered by the pyflare binary, which is effectively a Python interpreter just as the familiar python binary, slightly modified to play well with the rest of the Flare ecosystem (Figure 2).
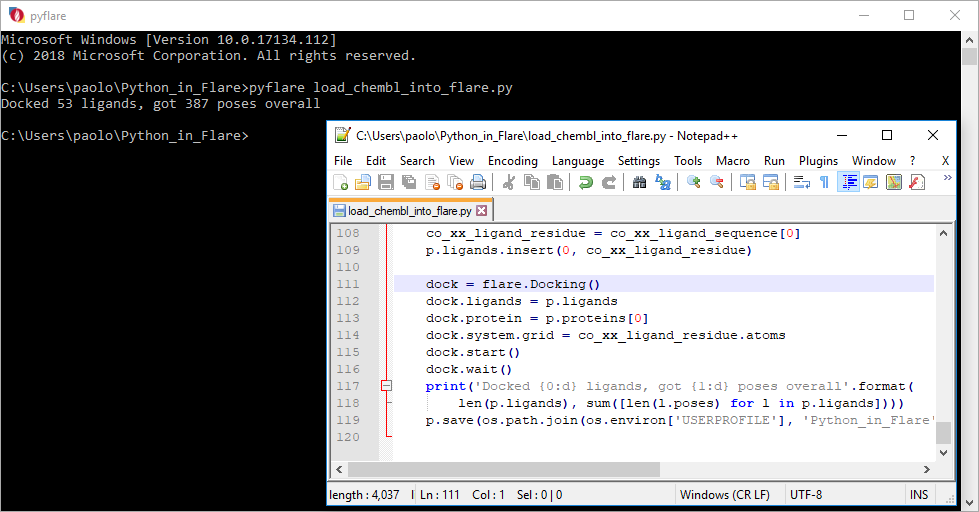
If your goal is to run a simple Python one-liner to, for example, list all cysteine residues in a protein chain or to print the distance between a ligand atom and a couple of key residues in the active site, the embedded Python Console is probably the simplest and leanest option – run the command and examine its output in the text port (Figure 3).
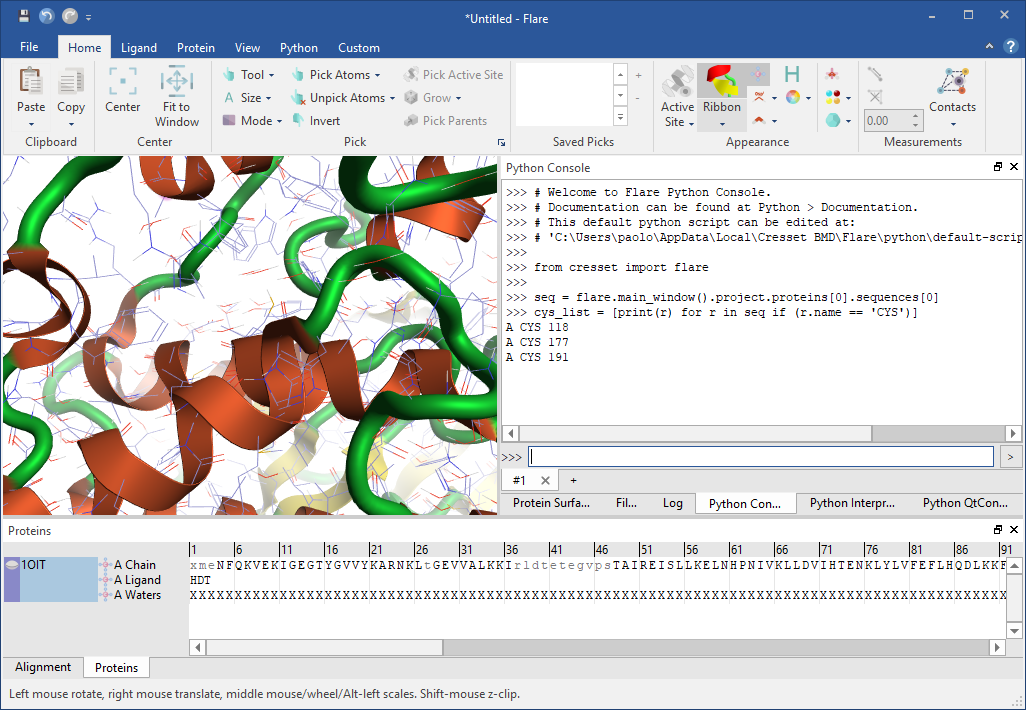
If you believe you will need to go through some iterations of a code-run-test-debug workflow on a somewhat more complex script, you will most likely choose the embedded Python Interpreter widget, which allows you to load a script, interactively edit it inside Flare and then save your modifications (Figure 4). Both the Python Console and the Interpreter come with a multi-tabbed interface that makes it possible to work on multiple Python snippets at the same time.
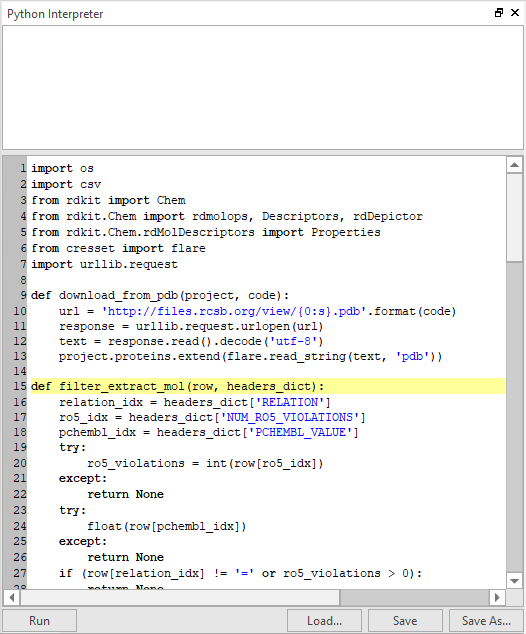
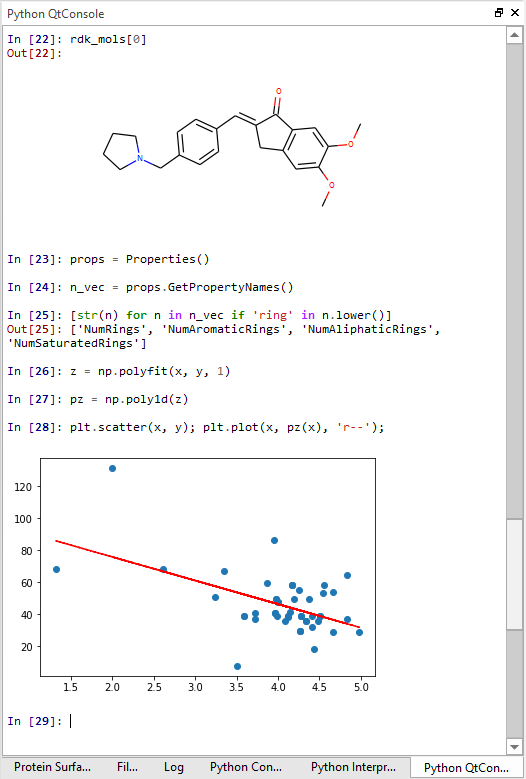
If you are used to the highly interactive environment of Jupyter notebooks, then you are going to love the Python QtConsole embedded in Flare. This widget provides all the nifty Jupyter features, i.e., TAB completion, auto-indentation, syntax highlighting, context help, inline graphics, and more (Figure 5). Type Python commands, examine molecules, draw plots – all in the same window.
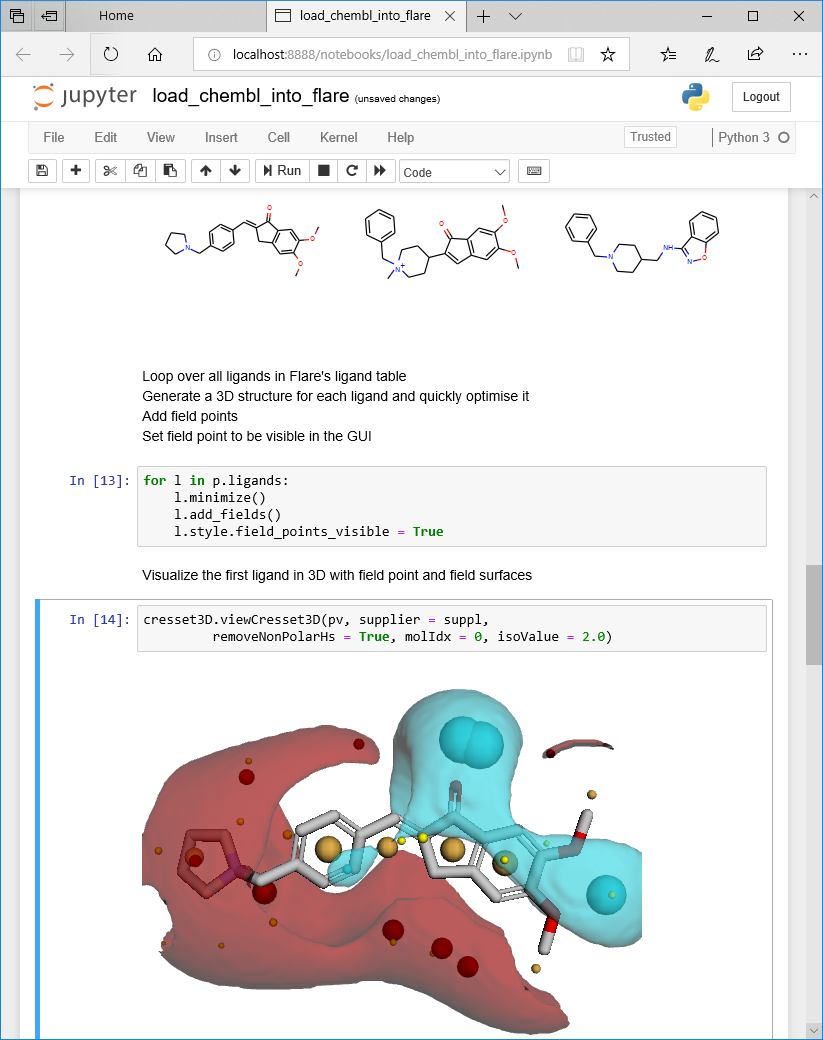
If you would rather prefer to use Jupyter Notebooks in the traditional way, i.e., multiple tabbed sessions inside your favorite browser, that’s entirely possible, too. You will even be able to look at ligands and proteins in 3D inside your notebook, including Cresset surfaces and field points (Figure 6).
In summary, programmatic access to Flare methods is very appealing to computational chemists looking for ways to automate tasks or run complex workflows on multiple datasets. On the other hand, the possibility to extend the core functionality as well as the graphical user interface with own methods makes Flare the ideal playground for Python developers.
For academic researchers, Flare is an opportunity to make their science more broadly accessible through integration into a user-friendly environment, while corporate users will appreciate the possibility to augment Flare capabilities with in-house REST services and tools. The possibility to code in Python taking advantage of the highly interactive experience provided by the Jupyter QtConsole and Notebook is a further incentive to make Flare the environment of choice for computational chemistry workflows.