Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Flare™, our most feature-packed software platform, receives bi-annual updates in response to customer feedback and to provide users with access to the very latest computational chemistry methods. With our eagerly anticipated spring release not too far away, the software development team is currently adding their finishing touches to what will become version 7 of the platform.
In anticipation of the release, let’s have a look together at some of the new features and improvements that will become available within Cresset’s ligand-based and structure-based molecular modeling platform.
In Flare V7 you will be able to run multiple jobs in parallel. There’s no need to wait for one job to complete before you start a new one.
For example, you’ll be able to download and prepare a list of proteins from the RCSB, then download an additional collection without having to wait for the first protein preparation to complete. Similarly, you will be able to initiate multiple docking or alignment experiments at once, each using slightly different parameters, which will be queued and processed as soon as local or remote resources become available.
An intelligent locking system prevents you from unintentionally deleting or modifying the ligands and proteins used in running jobs until they are finished. Furthermore, a re-designed calculations window (Figure 1) monitors all running, queued, and finished jobs, giving you also easy access to the Log for each calculation.
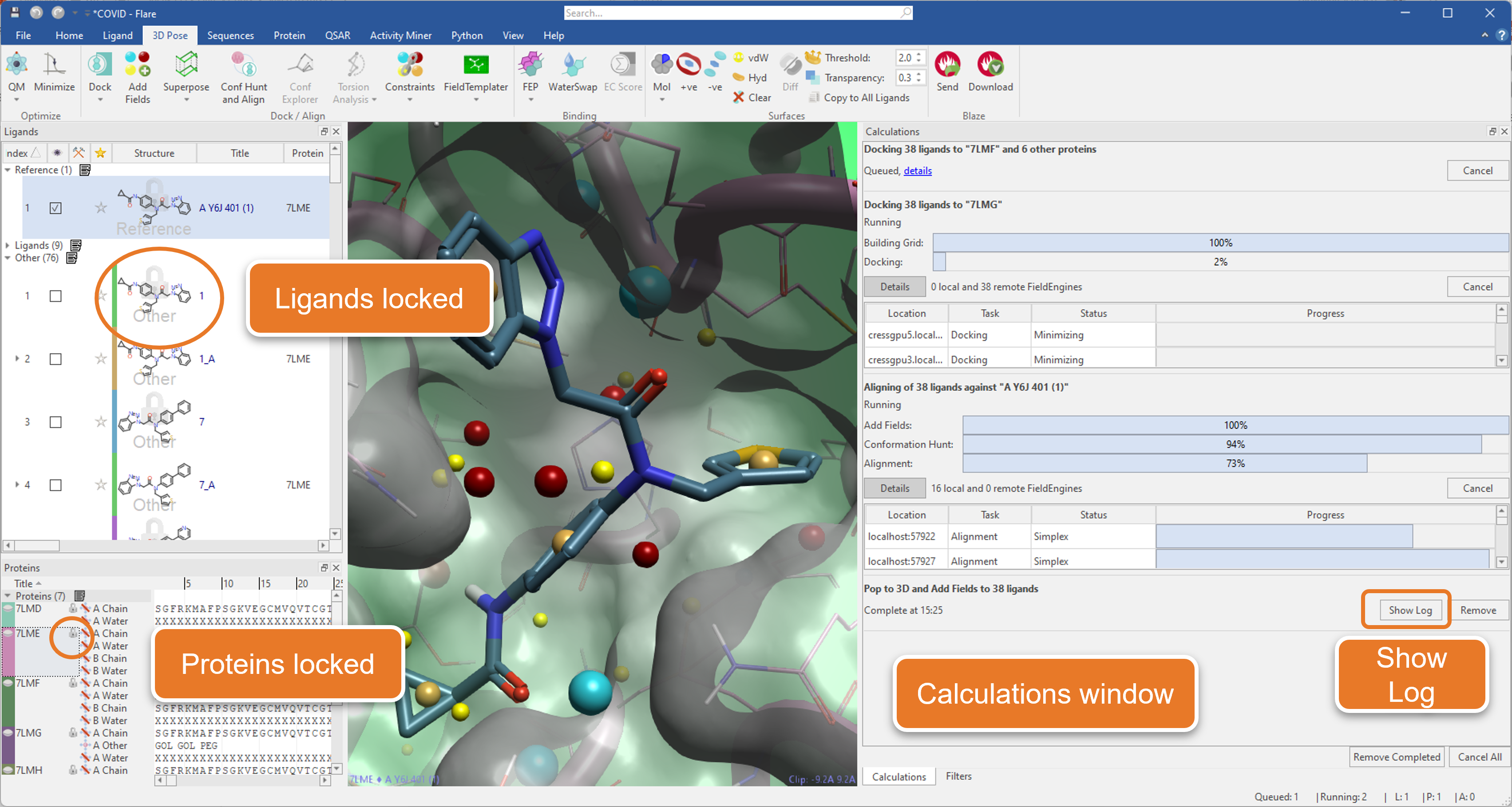
Figure 1. The new ‘Calculations’ window for monitoring multiple jobs in Flare.
In Flare V7 we have further expanded the choice of available docking workflows by merging two popular methods (ensemble and covalent docking) to enable Ensemble Covalent Docking calculations (Figure 2).
You can use this new method whenever you want to consider protein flexibility in your covalent docking experiment, by including multiple, alternative active site conformations of the same protein in a single covalent docking run. These can be experimentally derived or gathered from alternative sources, such as a molecular dynamics study.
In the example shown in Figure 2, the 2D structure of Branebrutinib with its alkynamide covalent warhead (orange circle) was docked into three different conformations of the active site of Bruton Tyrosine Kinase (BTK) bound to covalent inhibitors Ibrutinib (PDB: 5PJ9), Zanubrutinib (PDB: 6J6M), and Evobrutinib (PDB: 6OMU). Before starting the experiment, the three protein structures were carefully prepared and superimposed in Flare.
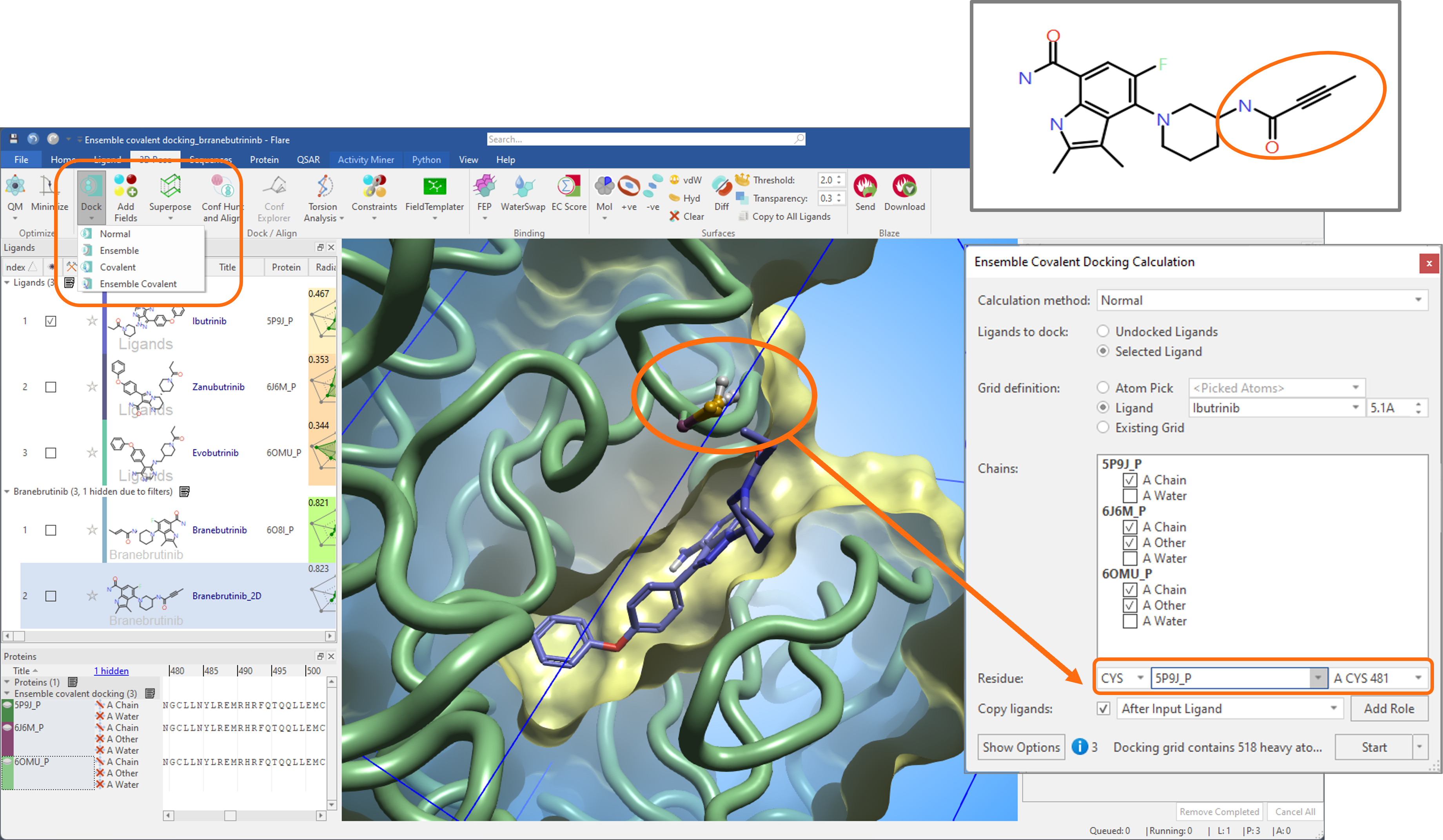
Figure 2. Ensemble covalent docking of Branebrutinib into three different conformations of the active site of BTK.
The 3D structure of Ibrutinib bound to PDB: 5PJ9 was used to define the docking grid, and CYS 481 in the prepared 5PJ9 protein structure was chosen as the covalent residue. Equivalent covalent residues in the superimposed 6OMU and 6J6M structures are automatically detected by Flare.
The results (Figure 3) show that the ensemble covalent docking workflow is capable to identify a pose for docked Branebrutinib (green) which closely mimics the crystallographic pose from PDB: 6O8I (purple) of this covalent inhibitor of BTK.
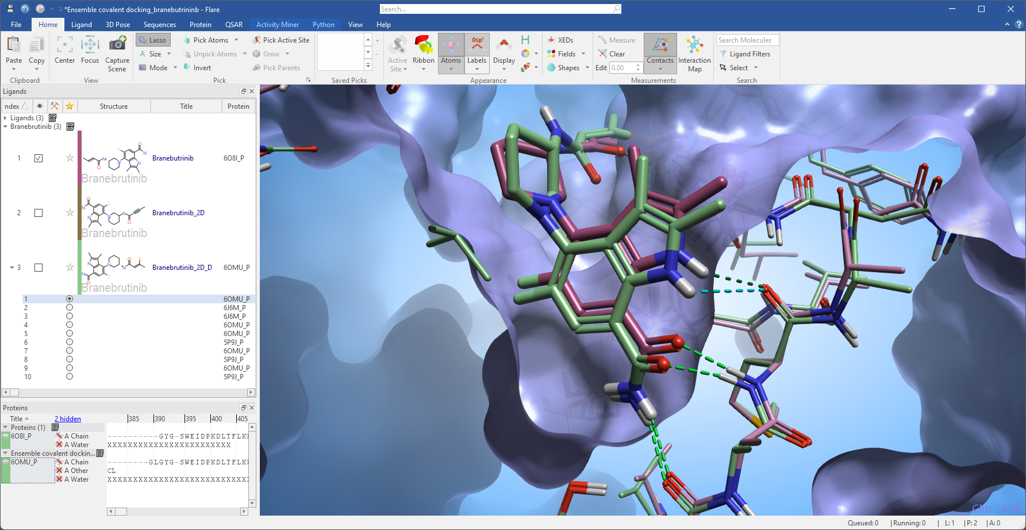
Figure 3. The ensemble covalent docking workflow identifies a pose for Branebrutinib docked in 6OMU_P (green ligand and protein) which closely mimics the crystallographic pose from PDB: 6O8I (purple ligand and protein) of this covalent inhibitor of BTK.
Additionally, for all docking methods in Flare it is now possible to specify which H-bond donor protein residues in the active site (choosing from each or all of Serine, Threonine, Tyrosine and Lysine) are allowed to rotate during the docking experiment (Figure 4). This provides an additional way of allowing side chain flexibility in your docking experiment and achieving optimal interactions with the protein active site.
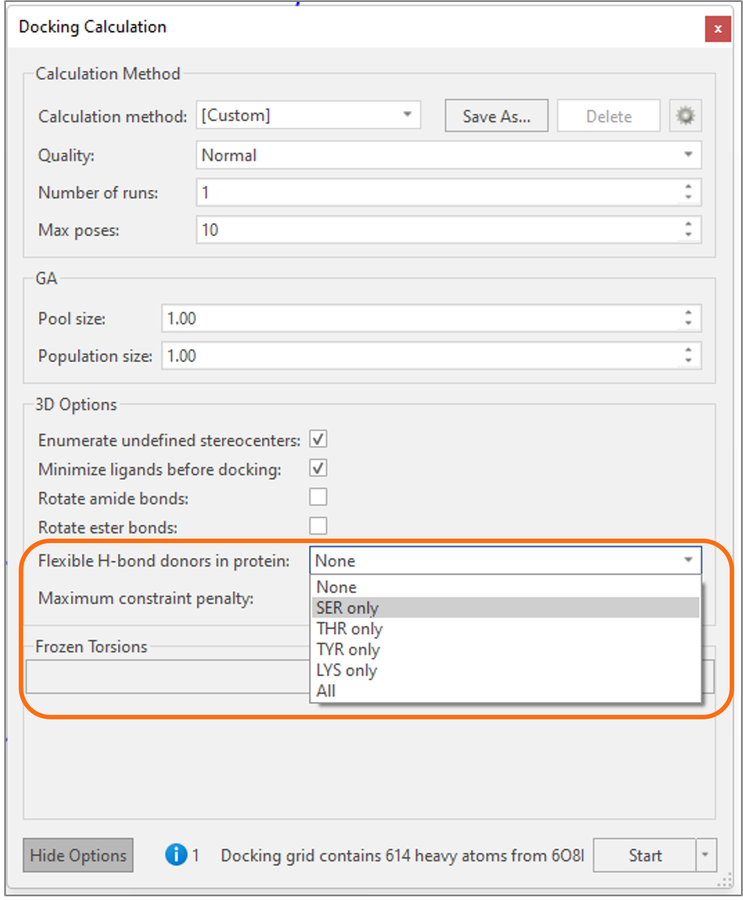
Figure 4. H-bond donor groups for Serine, Threonine, Tyrosine, and Lysine are allowed to rotate during the docking experiment by setting the desired ‘Flexible H-bond donors in protein’ advanced option.
Flare V7 features expanded capabilities and enhancements for building quantitative SAR models to robustly predict the activity and ADMET properties of your ligands.
A new ‘Consensus’ model type has been added to the available Regression and Classification models. Running a Consensus regression model (Figure 5 – left) will cause Flare to run all available machine learning regression models (Gaussian Process, MultiLayer Perceptron, Random Forest and Support Vector Machine) and predict the activity of compounds as the average of the predictions from each individual model. Consensus classification (Figure 5 – right) instead predicts the class/category of molecules as the most frequently predicted category from each individual machine learning classification model (MLP, RF and SVM).
This creates a ‘consensus’ for the predictions, enabling you to prioritize the ligands which all the regression or classification models agree would be a good idea to make.
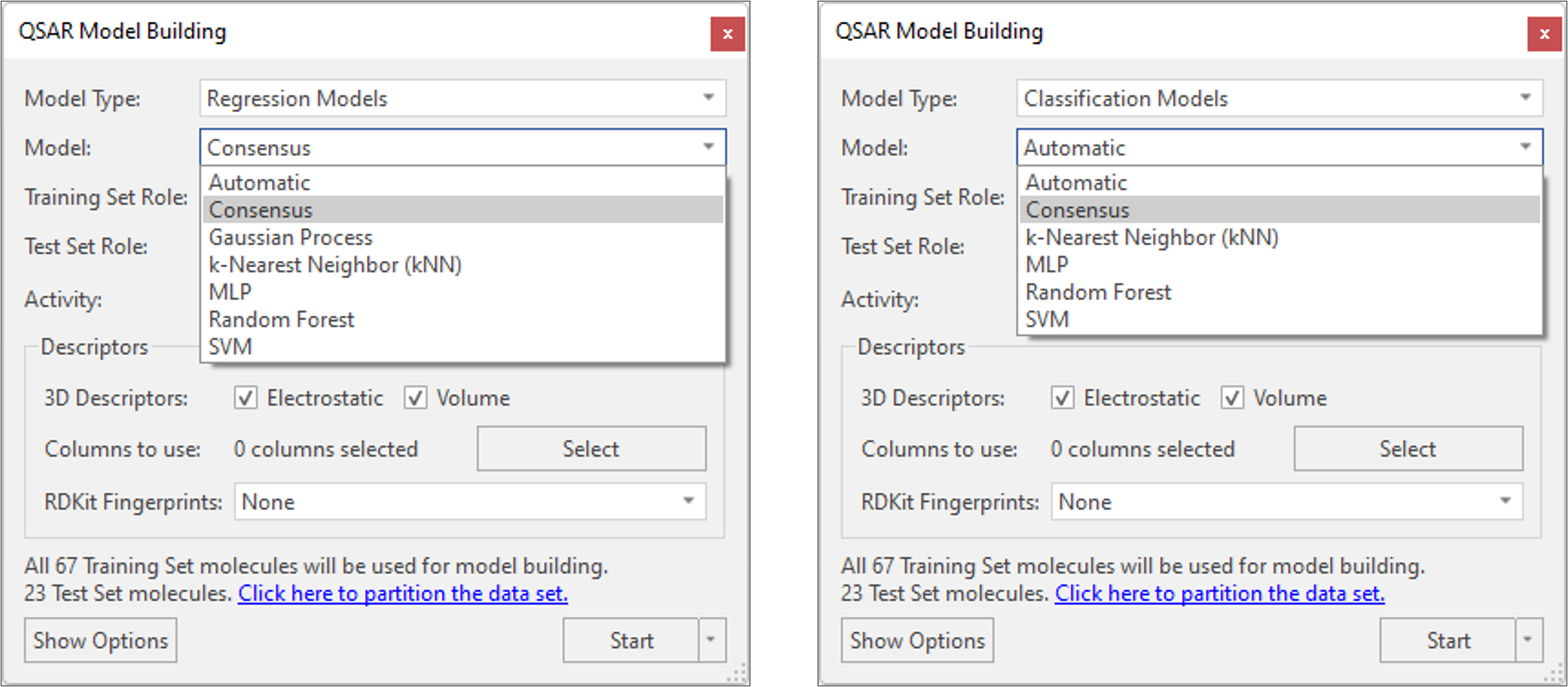
Figure 5. Consensus regression (left) and classification (right) models enable you to prioritize the ligands which all the machine learning regression or classification models agree would be a good idea to make.
Within Flare V7, we have further streamlined the use of additional physico-chemical descriptors from the RDKit for building predictive QSAR models of activity and ADMET properties, as an alternative to Cresset 3D descriptors of Electrostatic and Shape.
Both regression and classification models can be run seamlessly on a choice of different fingerprints, while several widely used physico-chemical descriptors can be imported into the Ligands table from the Column & Activity Editor (Figure 6).
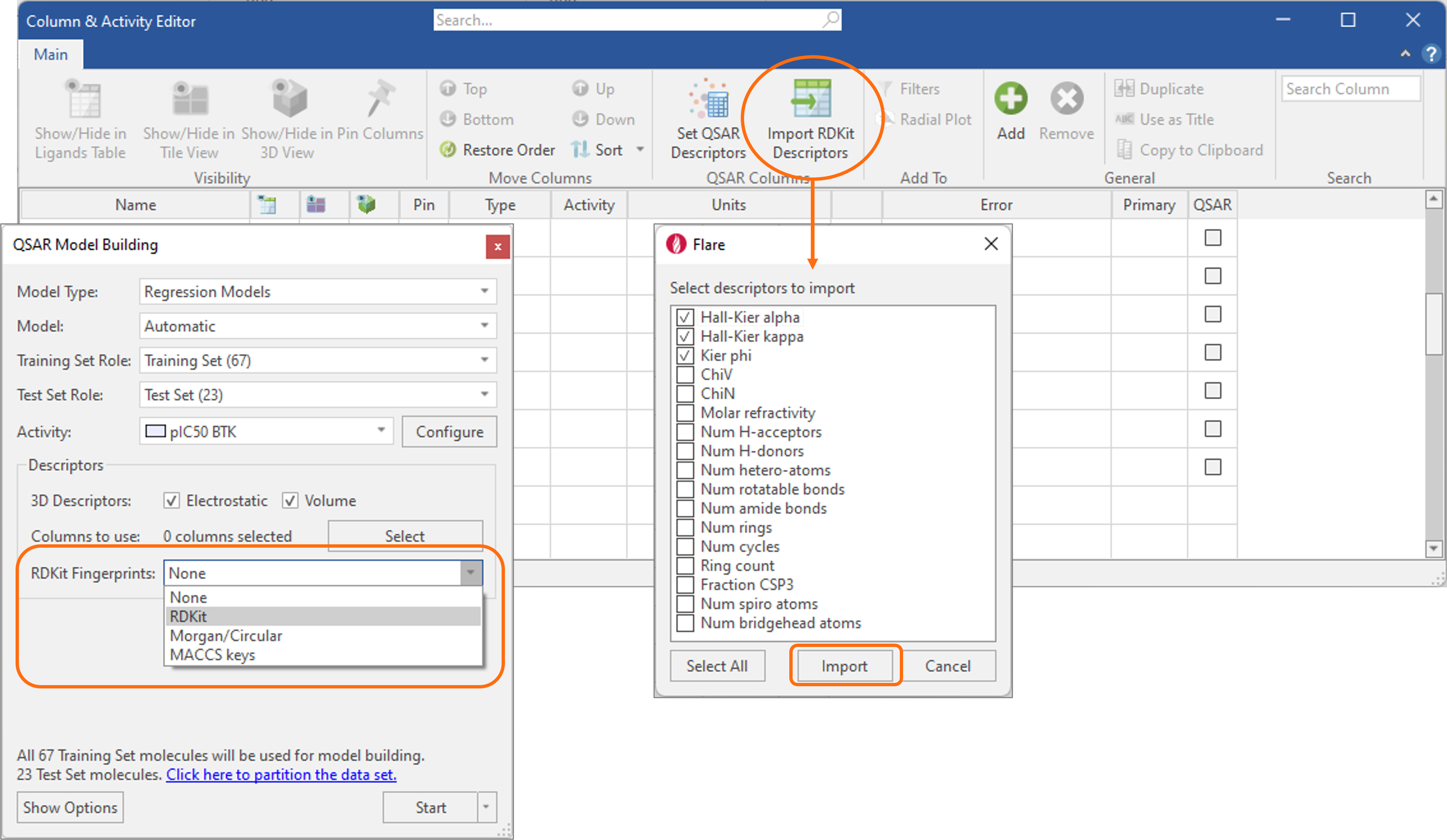
Figure 6. RDKit fingerprints and descriptors can be used in Flare to build predictive QSAR models of activity and ADMET properties.
Finally, Gaussian Process regression models now calculate standard deviation values for each predicted value (Figure 7). Typically, predictions associated with standard deviation values much higher than those for the training set used to build the model are less accurate, enabling you to gain confidence in the reliability of the predictions made by Flare before the compounds are synthesized and tested.
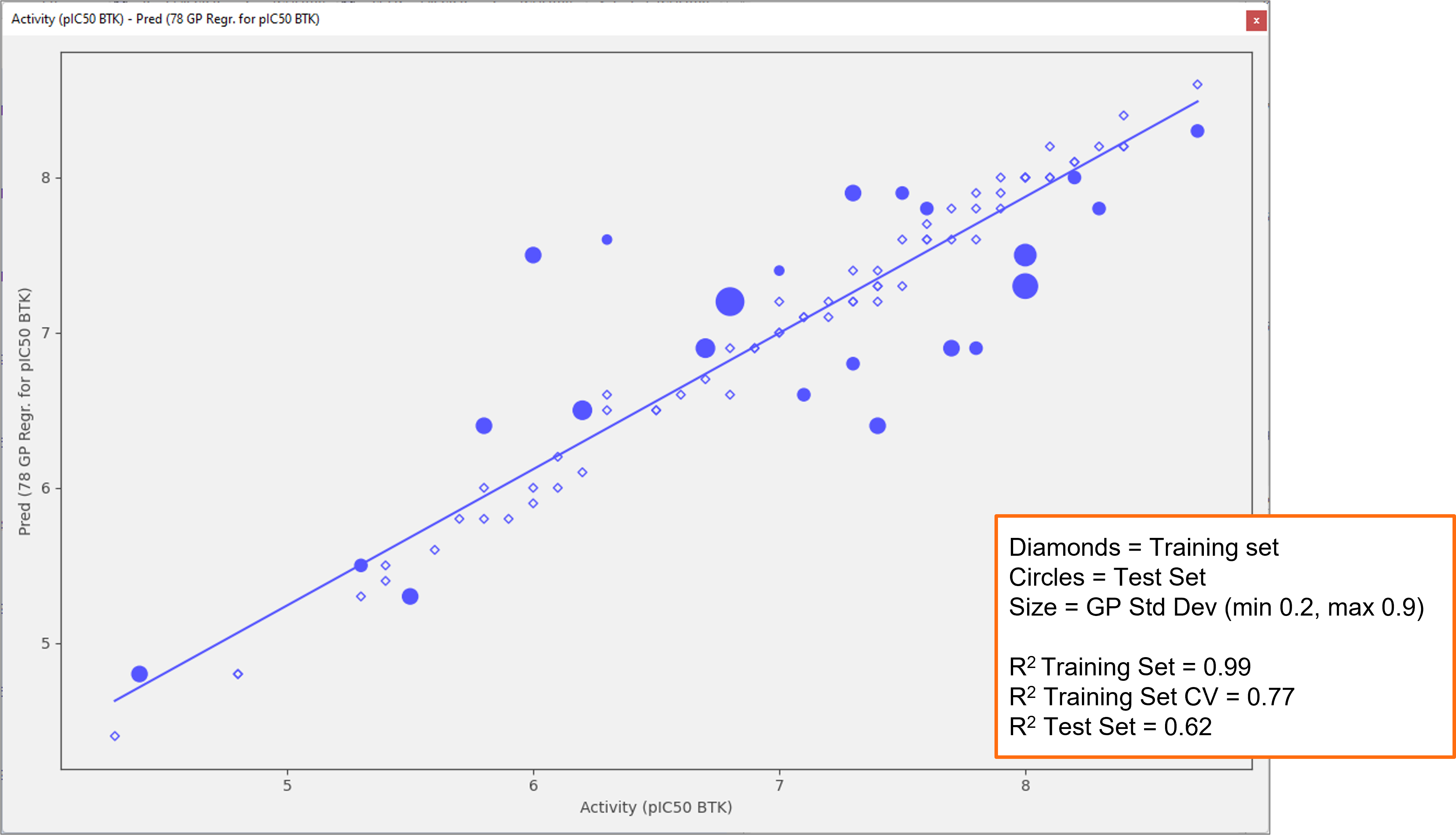
Figure 7. Gaussian Process models in Flare V7 calculate a standard deviation value for each prediction, enabling you to gain confidence on the reliability of the predictions before the compounds are synthesized and tested.
Water molecules directly bound to proteins often exchange slowly with bulk solvent, making some systems very slow to equilibrate during molecular dynamics simulations. In these situations, grand canonical methods such as Grand Canonical Monte Carlo1 (GCMC) can help accelerate the equilibration process by allowing the number of water molecules in a simulation to fluctuate. This is achieved by performing Monte Carlo moves that insert and delete water molecules into/from the active site, allowing an accelerated exchange of waters with bulk water.
GCNCMC2 combines traditional GCMC with Nonequilibrium Candidate Monte Carlo (NCMC) into Grand Canonical Nonequilibrium Candidate Monte Carlo (GCNCMC), in which water insertions and deletions are carried out in a gradual, nonequilibrium fashion, improving the efficiency of water sampling and often leading to increased sampling of bound ligand conformations.
More efficient water sampling with GCNCMC will be available in Flare V7 for both molecular dynamics and Free Energy Perturbation (FEP) experiments. For molecular dynamics, GCNCMC can be used over the whole duration of the simulation or during the equilibration stage only (Figure 8). For FEP, GCNCMC can be used during the equilibration stage.
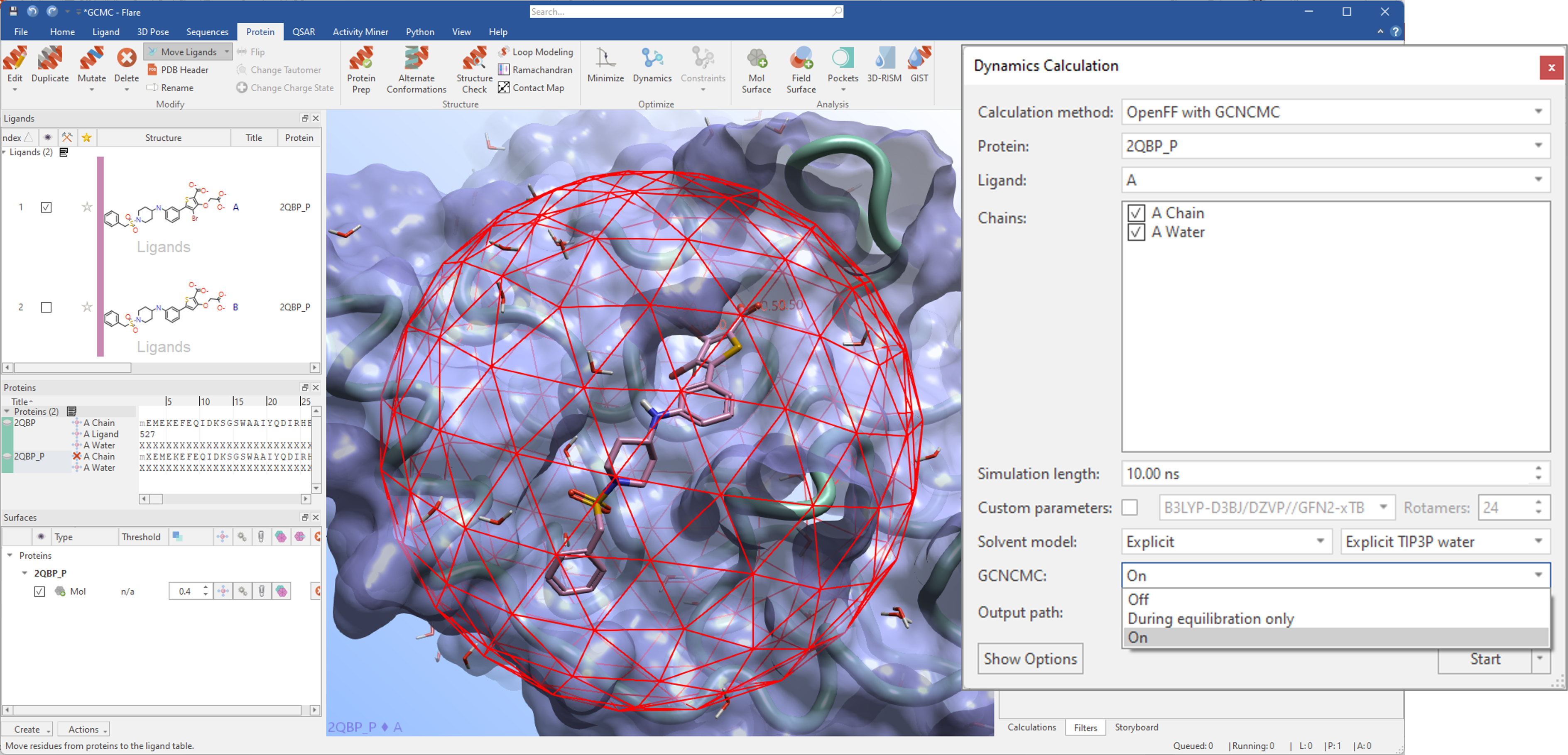
Figure 8. GCNCMC can be used in molecular dynamics experiments to improve water sampling and sampling of bound ligand conformations during the equilibration and production stages of the simulation.
Using GCNCMC during the equilibration stage of Flare FEP calculations can significantly boost the predictivity of the method, especially in protein-ligand complexes where the active site features occluded pockets, typically difficult to hydrate.
The example shown in Figures 9 and 10 is taken from our validation experiments. The two PTP1B inhibitors in Figure 9 differ only for the presence/absence of a bromo substituent on the thiophene ring (orange circle), resulting in a difference in the ΔG of binding equal to 3.6 kcal/mol.3
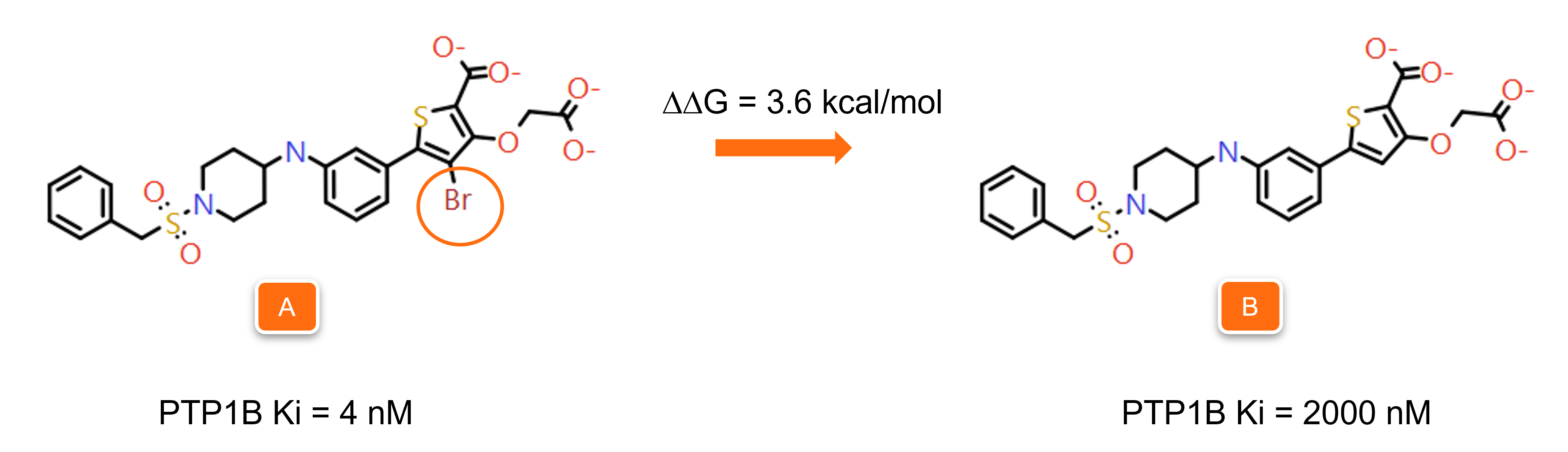
Figure 9. Molecules A and B differ only for the presence/absence of a bromo substituent on the thiophene ring, resulting in a difference in the ΔG of binding of 3.6 kcal/mol.
In Figure 10 we can see the results of Flare FEP calculations ran on PDB: 2QBP in default conditions (left) and using GCNCMC (right). Using GCNCMC during the equilibration stage of the simulation significantly improves the hydration of the active site, leading to more accurate Flare FEP predictions.
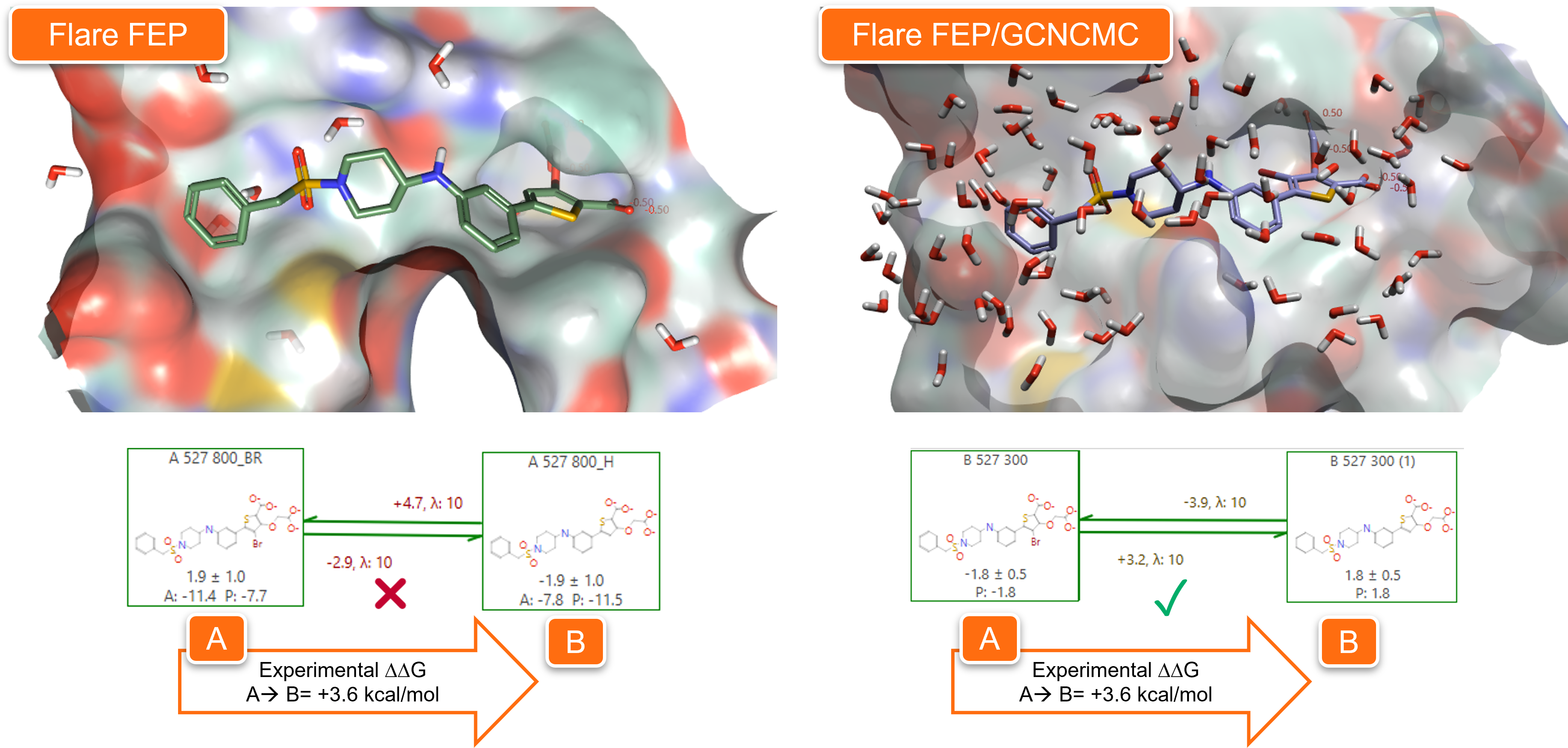
Figure 10. Using GCNCMC during the equilibration stage of Flare FEP calculations to transform ligand A (from PDB:2QBP) into ligand B significantly improves the hydration of the active site and increases the accuracy of the FEP predictions.
The next release of Flare also includes several new features and improvements to make the GUI more appealing and easier to use. Two key additions are the ability to search for a Flare feature (Figure 11) and the new ‘Pin Columns’ functionality (Figure 12).
The ‘Search’ box at the top of the Flare window is useful when you want to quickly locate a Flare item, whether it is a button, a menu item, or a dockable window. For example, a user might want to set docking constraints of a protein residue but isn’t sure where to find this feature. Typing ‘dock’ into the search box will offer a choice of options matching the search criteria (Figure 11). From the list, ‘Constraints’ then ‘Add Docking Constraints to Protein Atom(s)’ can be chosen, and this will automatically open the desired menu.
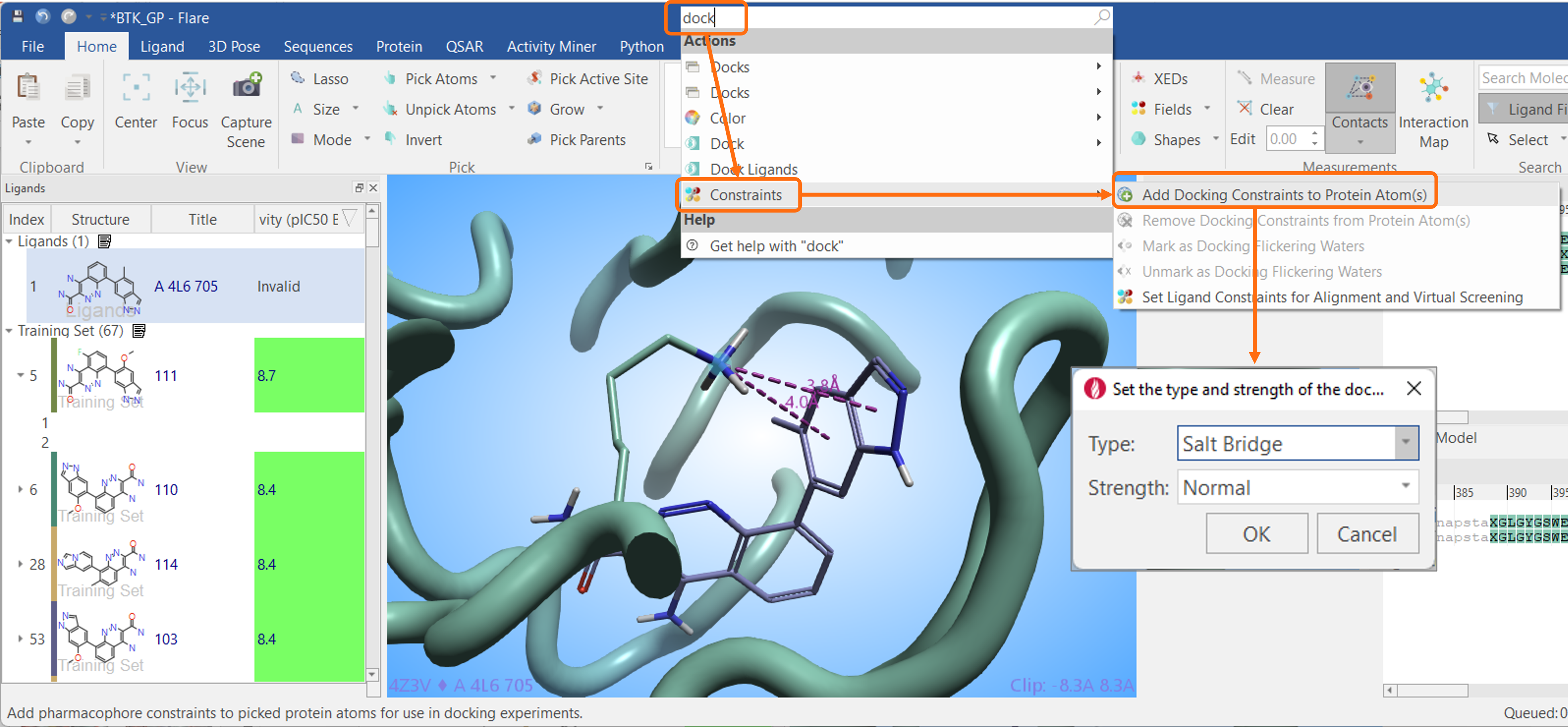
Figure 11: Use the ‘Search’ box at the top of the Flare window to rapidly find the desired functionality in Flare.
Pinning columns enables you to keep one or more columns always visible, or ‘pinned’ to the left side of the Ligands table. In the example shown below, the ‘Structure’, ‘Title’ and ‘Activity’ columns are selected so that they are always visible when scrolling the Ligands table to the right to explore other properties. The ‘Pin’ function is conveniently available from both the right-click menu of most Ligands table columns, and from the Columns & Activity Editor (Figure 12).
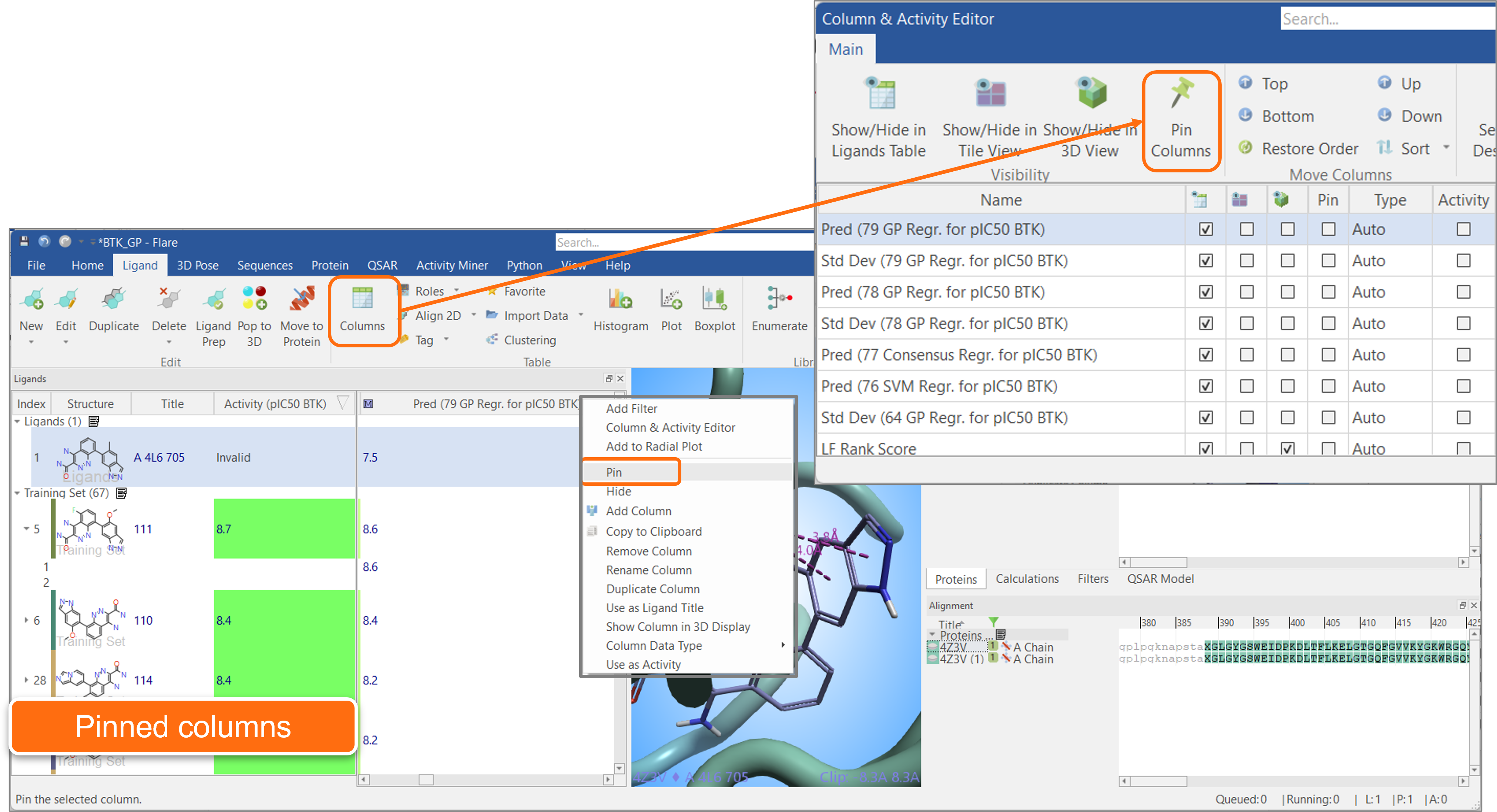
Figure 12. ‘Pin’ your favorite columns to keep them always visible on the left side of the Ligands table.
As we hope you’ll be excited to see, the Spring 2023 release of Flare, Cresset’s ligand-based and structure-based molecular modelling platform, is packed full of new science, new features, and usability improvements. There’s not long to go, so we’ll let you know as soon as the updates are finalized, and the new version of the platform becomes available.
Stay tuned for our release announcement due next month, and an exciting webinar, planned for June, where we’ll be demonstrating and exploring the new features in more depth.