Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
One of the many new features in Flare™ V6 is Torsion Analysis, which enables visual identification of problematic torsions on the fly. Torsion Analysis facilitates novel molecule design, by monitoring the quality of the torsions of ligand conformations and poses, to give an indication of which torsions are most frequently seen experimentally. The remarkable usability of this method is also attributed to the fact that it can be used not only on ligand conformations and poses, but also during ligand editing to determine the torsional quality while designing new molecules.
The method works by updating the color of the atoms involved in the torsion, depending on the frequency of finding the specific torsion in the Cambridge Structure Database (CSD).1 Torsion Analysis in Flare is based on the Torsion Library method,2,3 which analyzes the content from databases such as the CSD. Molecular design experts have curated hundreds of rules for small molecule conformations, encoded by SMARTS, categorizing torsions using a traffic light coloring scheme. Depending on the frequency that each torsion is found at the CSD, green represents torsions of high frequency, yellow for the medium frequency and red for the low frequency ones.
We will use two Protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK) inhibitors with a common indoline core4 to demonstrate this functionality. The molecules in this dataset are highly congeneric, presenting only peripherical group variations to a common core. From this dataset, compound 38 (GSK2606414) is a potent and selective PERK inhibitor and can be found bound to the protein in crystal structure PDB: 4G31.
A 3D conformation for compound 1 (Figure 1a) was created using the ‘Pop to 3D’ button in Flare. The Torsion Analysis method was applied to this conformation and two low frequency torsions (in red) were identified with the help of the coloring scheme. This is a possible concern and suggests the molecule should be edited to generate torsions which are found more frequently, and create a more realistic 3D conformation for this ligand.
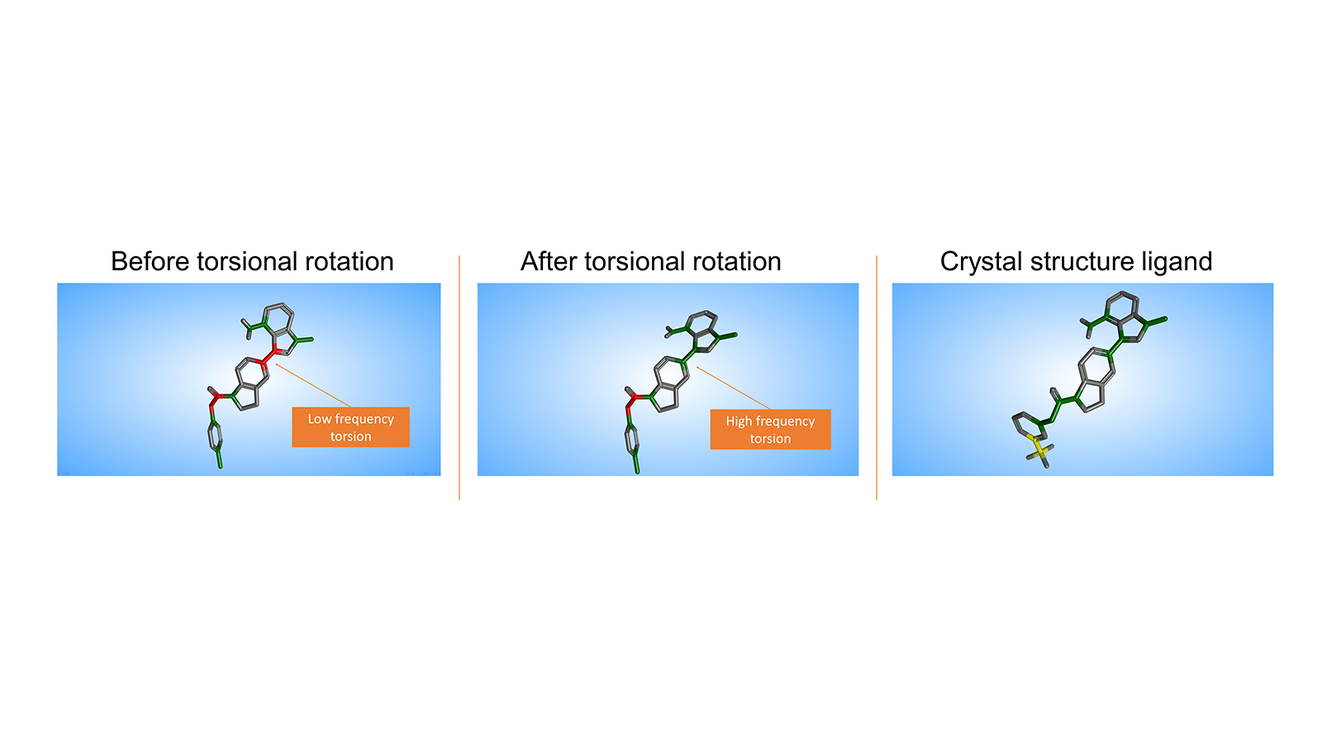
Figure 1: Left: Compound 1 before torsional rotation. The low frequency torsions are colored red; Centre: Compound 1 after torsional rotation - the previously problematic torsion is now colored green, as it is found very frequently in the CSD; Right: Crystal structure of compound 38. The torsion is similar to the one of compound 1 after the torsional rotation.
This can be easily done using Flare’s editing platform: while rotating the bond, the energy profile is shown in real time as shown below. This profile informs us whether the torsion sits on the low, medium or high frequency region of the CSD.
After rotating one of the problematic bonds, the Torsion Analysis method can be run again to confirm that the torsion is indeed of high frequency (green, Figure 1b). Comparison with the crystallographic ligand (Figure 1c) shows that this torsion is indeed preferred for binding, as it is similar to the torsion adopted by compound 38 at the crystal structure.
The Torsion Analysis method also reports the number of torsions with a high, medium and low frequency for each ligand and it can be used on conformation ensembles, as well as on poses created with Docking and Conformation Hunt and Align experiments.
See for yourself how intuitive Torsion Analysis is for enhancing novel molecule design in your project – request an evaluation of Flare.