Jessica Plescia†, Stuart Firth-Clark†
†Cresset, New Cambridge House, Bassingbourn Road, Litlington, Cambridgeshire, SG8 0SS, UK.
Abstract
Predictive 3D quantitative structure-activity relationship (QSAR) methods for hit and lead optimization are invaluable in early drug discovery stages. In this case study, the computer-aiding drug design (CADD) platform Flare1 was used to demonstrate the predictive capabilities and advantages of our 3D QSAR in building predictive models of phosphodiesterase 10A (PDE10A) inhibitor activity. Utilizing a dataset of 1162 ligands and 77 crystal structures of PDE10A, we demonstrated that not only can Flare’s 3D ligand alignment tool be reliably applied to diverse data sets, but that 3D QSAR based on ligand alignment exhibited more robust results than using molecular docking and does so on a shorter time scale. This method successfully demonstrated that ligand-based methods are more than a viable option in the analysis of large, diverse data sets.
Introduction
Molecular docking is a powerful predictive tool and one of the most common structure-based methods in drug discovery. With reliable protein structural information, it can be an extremely useful method, especially when used in conjunction with powerful ligand-based methods (e.g., virtual screening combined with ligand-based 3D property scoring) However, there are many factors to consider, including protein conformations and flexibility.2 Furthermore, a well-modeled crystal structure of the target protein is not always available, creating a need for reliable ligand-based methods. Fortunately, for such scenarios, there exist ligand-based methods, such as ligand-based virtual screening and qualitative or quantitative structure-activity relationships (QSAR).
Flare’s 3D QSAR methods require reliable alignment of ligand structures based on shape and electrostatics. Once the ligands have been conformationally hunted, Flare uses Cresset field points to identify positive and negative electrostatics, as well as van der Walls surface (shape) and hydrophobicity to align the ligands. With these field points, the software creates a model pattern of the ligands in terms of how the protein target sees them – by electrostatics and shape – without the need for a protein structure.3 Figure 1 shows the fields and field points for Mardepodect, a PDE10A inhibitor used to treat schizophrenia.4
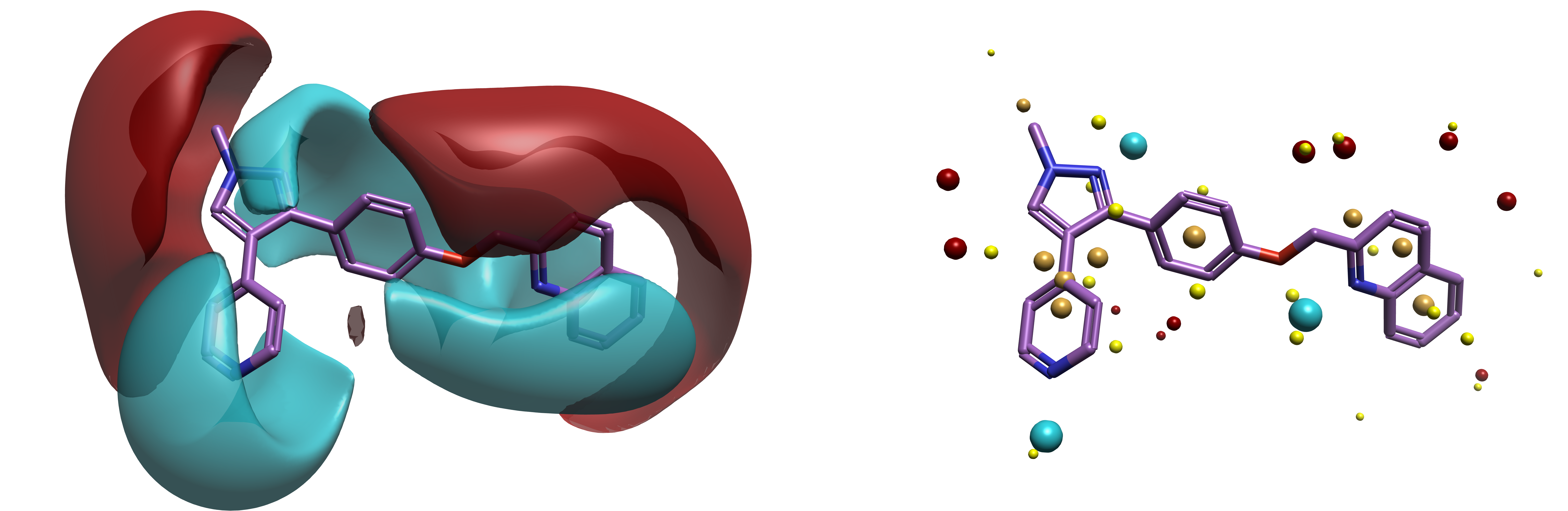
Figure 1. Bioactive conformation of PDE10A inhibitor, Mardepodect (pdb 3HR1).3 (Left) Shown with Cresset fields. Red = positive, blue = negative. (Right) Shown with Cresset field points. Red = positive, blue = negative, yellow = van der Waals shape, gold = hydrophobicity.
For this case study, the dataset from Tosstorff et al.5 was used to demonstrate the predictive capabilities of Flare’s 3D QSAR methods. This dataset consisted of 77 crystal structures of PDE10A, and their associated co-crystallized inhibitors, plus an additional set of 1162 inhibitors with activity data, all assessed with the same biochemical assay. While the authors studied these inhibitors with multi-template molecular docking, this study will compare results from Flare’s 3D QSAR methods with Flare’s template docking.
Methods
Data preparation
Prior to any analysis of the ligand activity set, the data needed to be prepared for QSAR experiments in Flare. The following workflow was utilized to do so:
- Download the pdbs from the Protein Data Bank and prepare the proteins and ligands
- Align the amino acid sequences and superpose the crystal structures
- Categorize co-crystallized ligands by substructure
- Divide the co-crystallized ligands by substructure and align the inhibitors to them.
The first task was to prepare the 77 crystal structures to (1) extract the co-crystallized reference ligands in the bioactive conformations and (2) prepare the proteins to be used as excluded volume in subsequent experiments. To do this, the pdb codes were copied from the Supporting Information and the corresponding structures downloaded into Flare. Once they were imported, it was clear that there were ‘excess’ protein monomers that could be removed for simplicity. This was done using a snippet from the Flare Python Cookbook. With exclusively A-chains in the Proteins table, the proteins were then prepared, which included defining the active site and extracting the reference ligands. The activity data from the Supporting Information was then merged with the molecules in the Ligands table.
Next, the PDE10A crystal structures were sequence aligned and superimposed. Superimposing the proteins in Flare also results in corresponding crystallographic ligands being superimposed. Unsurprisingly, the sequence identities for the proteins were all > 98%, and the maximum RMSD was 0.55 Å. Figure 2 shows the crystal structure 5SDU and its eight most diverse crystal structures by RMSD. As is clear from the superimposition, they are still very close in 3D space.
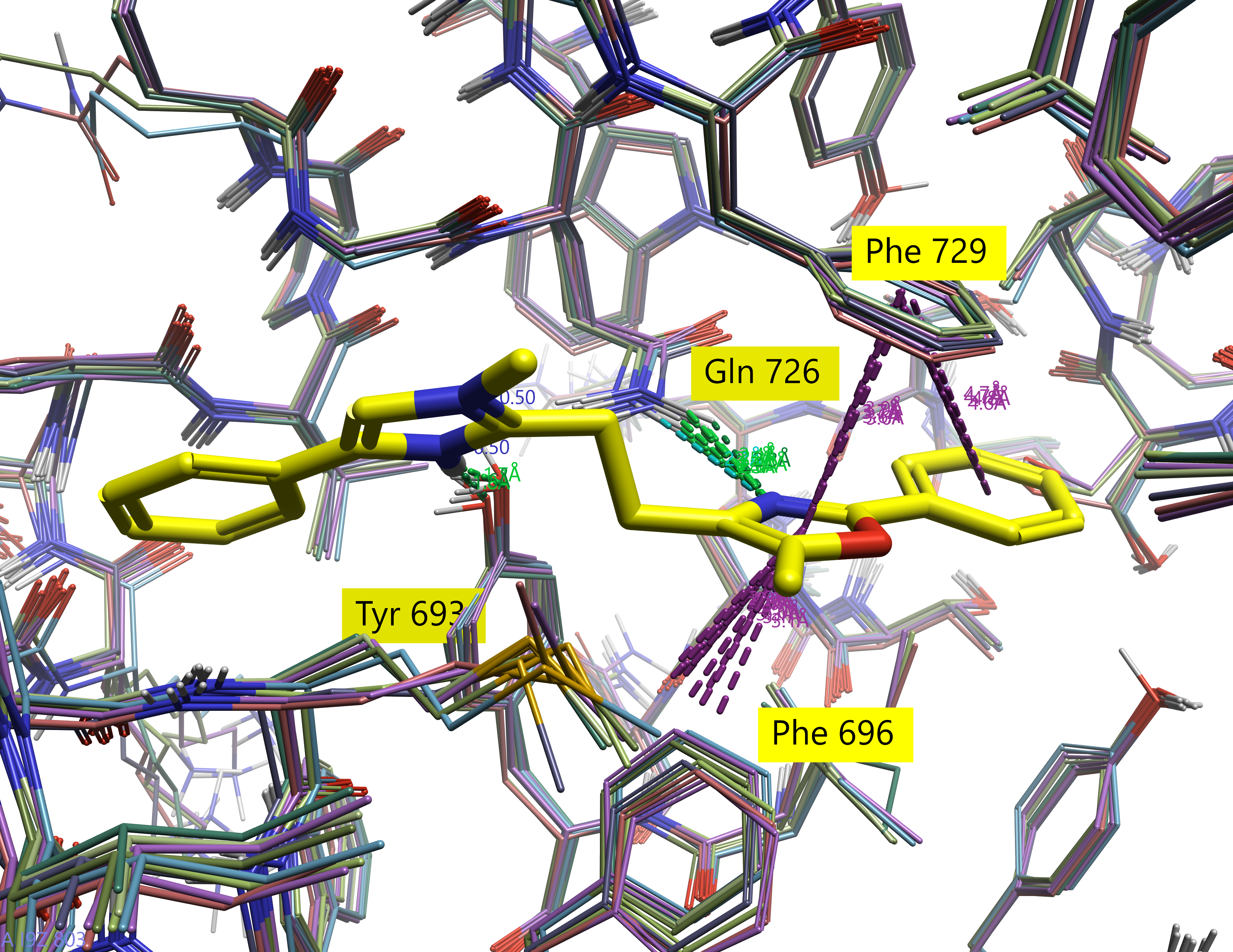
Figure 2. 5SDU and its reference ligand (yellow) superimposed to the eight most diverse crystal structures by RMSD: 5SDY, 5SE1, 5SEF, 5SEW, 5SEW, 5SF2, 5SFB, 5SFS.
As shown in Figure 2, the reference ligand makes key stabilizing interactions with each crystal structure, i.e., the positions of key residues for ligand stabilizing interactions are conserved across the crystal structures.
The 1162 ligands with their PDE10A activities were next imported and converted to 2D structures in Flare. They were previously divided into three classes of substructure by the authors, labelled aminohetaryl_c1_amide, aryl_c1_amide_c2_hetaryl, and c1_hetaryl_alkyl_c2_hetaryl. A representative ligand from each class is shown in Figure 3.
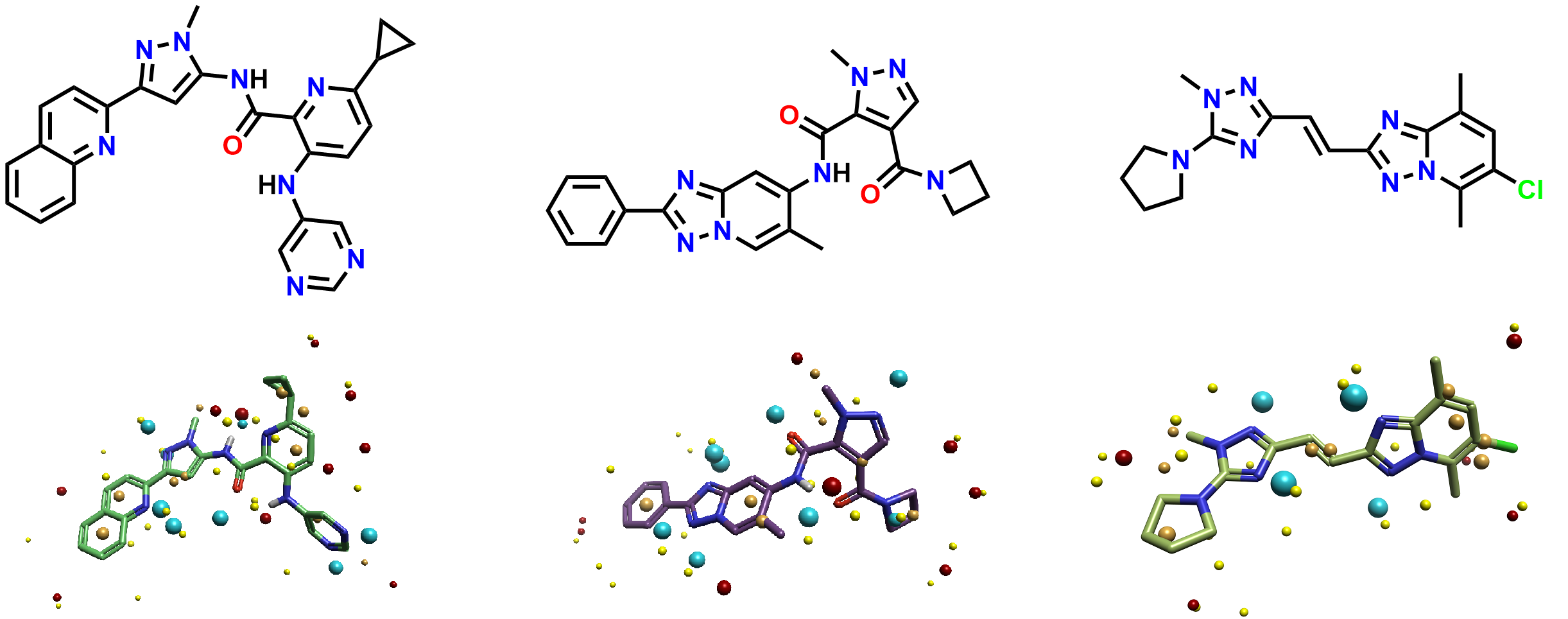
Figure 3. The most active ligand in each class and their respective Cresset field points (2D schematic representations shown above). (Left) aminohetaryl_c1_amide. (Middle) aryl_c1_amide_c2_hetaryl. (Right) c1_hetaryl_alkyl_c2_hetaryl.
Ligand-based technique: Maximum Common Substructure (MCS) ligand alignment for a QSAR study
In order to conduct QSAR experiments, each subset required alignments to the corresponding references. Choosing the best references is a very important step in a ligand alignment protocol; it is necessary to select references that are representative of the entire dataset in order to optimize the final alignment and hence the QSAR model accuracy. For example, choosing exclusively unsubstituted references, such as ligands with unsubstituted phenyl rings, would lead to ortho and meta-substituted ligands with substituents aligned in both orientations and therefore more noise in the final QSAR model. In this case, although most of the ligands in each dataset have very similar scaffolds, the R-groups varied greatly. Accordingly, nine references were chosen for each data subset to best represent the diversity of the ligand structures. Figure 4 illustrates this selection process. The 3D view shows four of the nine references that were chosen for the aminohetaryl_c1_amide subset.
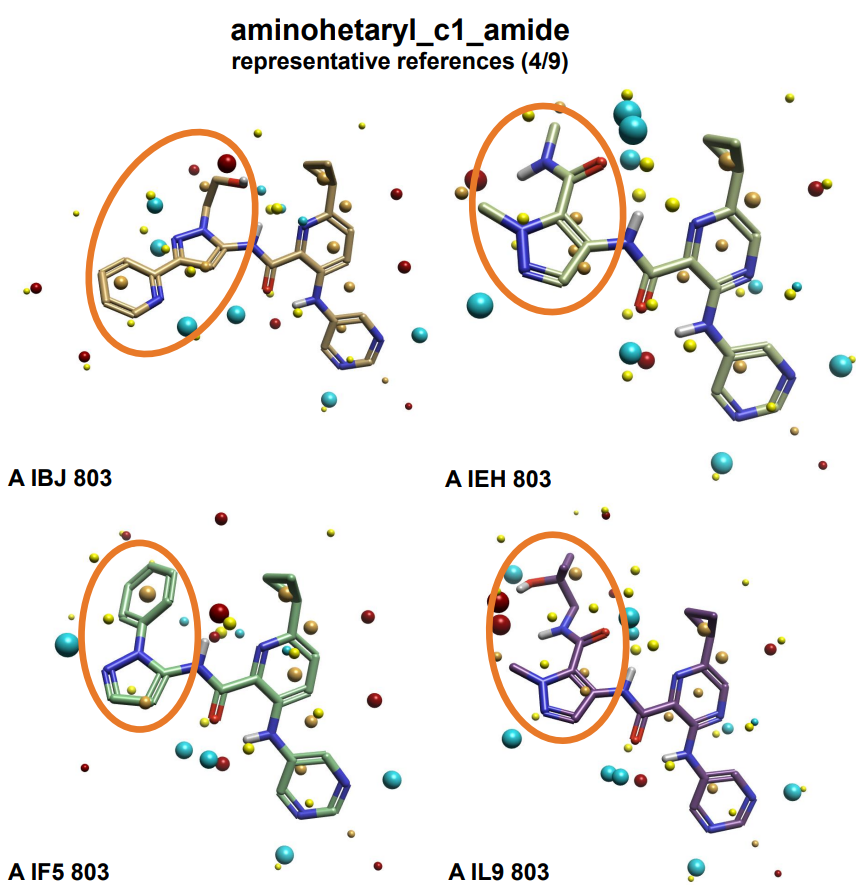
Figure 4. Four of the references for the aminohetaryl_c1_amide subset.
The four crystal structures have differently substituted pyrazoles on the western portion of the molecule. This selection provided several different reference field point patterns for shape and electrostatics, allowing differently substituted ligands in the aminohetaryl_c1_amide subset to be aligned to the closest match in the references. If, instead, A IF5 803 was chosen as the sole reference (Figure 4, bottom left), this might have resulted in unwanted rotations around the amide-pyrazole bond in the ligand alignment process. This process was repeated to choose representative references for each of the three ligand subsets.
Once nine representative reference ligands were chosen for each subset, the conformational hunt and alignment processes were run. Because of the size of the datasets and the flexibility of some of the ligands, the ‘Accurate but Slow’ setting was chosen for the conformational hunt to increase the total number of conformations and decrease the energy window cutoff. The alignment was then done using ‘MCS’ with ‘Permissive’ (hybridization matching) rules: MCS alignment first finds a commonality between each reference molecule and each ligand to be aligned in terms of MCS. The MCS region is frozen, and the rest of the molecule is conformationally hunted and then aligned to the reference using shape and electrostatics. Because we have access to protein structural information in this case study, the protein was factored in as an excluded volume, i.e., alignments that encroached into the active site were penalized. The pdb selected for excluded volume was that which was co-crystallized to the most active reference ligand. Figure 5 shows the alignments for each of the three subsets of ligands.
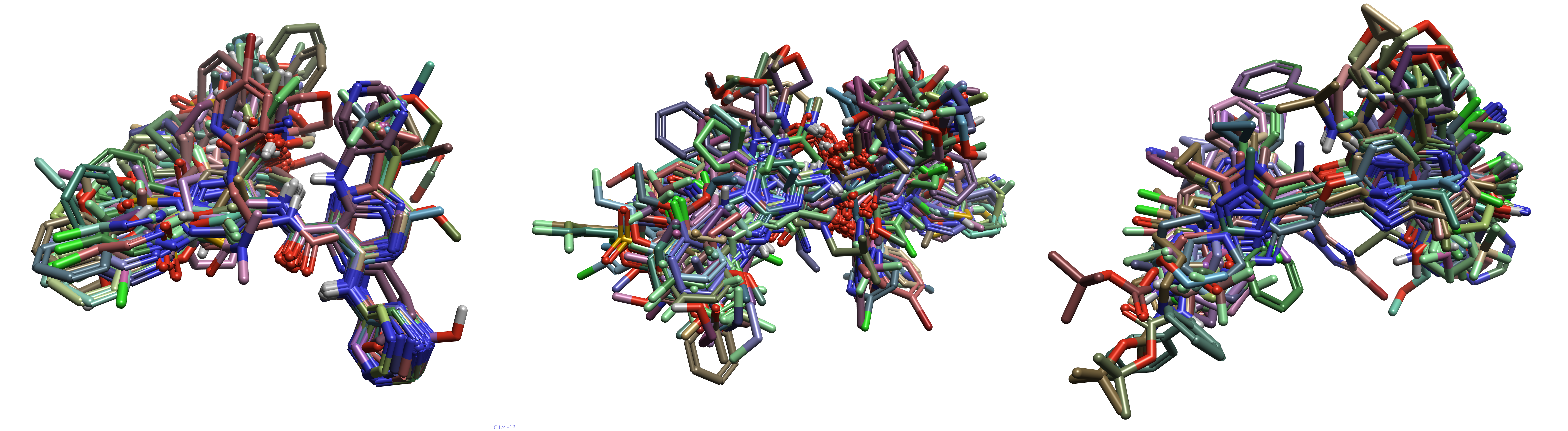 Figure 5. Ligand alignments of each subset. (Left) aminohetaryl_c1_amide, (Middle) aryl_c1_amide_c2_hetaryl, (Right) c1_hetaryl_alkyl_c2_hetaryl
Figure 5. Ligand alignments of each subset. (Left) aminohetaryl_c1_amide, (Middle) aryl_c1_amide_c2_hetaryl, (Right) c1_hetaryl_alkyl_c2_hetaryl
From Figure 5, it’s clear that while there is some commonality in the 2D substructure within each ligand set, they are still quite diverse. These alignments were used to build QSAR models for each ligand subset.
Ligand-based technique: electrostatic field- and shape-based ligand alignment for a QSAR study
The other type of alignment available in Flare is the ‘Normal’ alignment, using Cresset’s field-based alignment and scoring method, based solely on electrostatics and shape of the molecules to be aligned rather than chemical structure. This type of alignment is more biologically relevant, as it ignores the 2D structure and focuses only on 3D shape and electrostatics. It is also more convenient for diverse ligand structures, which we see when we look at the dataset of 1159 ligands as a whole.
To perform this screening, the three most potent reference ligands from each structure subset were used as references for a Normal alignment in Flare, using the Accurate but Slow conformation hunt. Figure 6 shows the overall ligand alignment of the 1159 ligands.
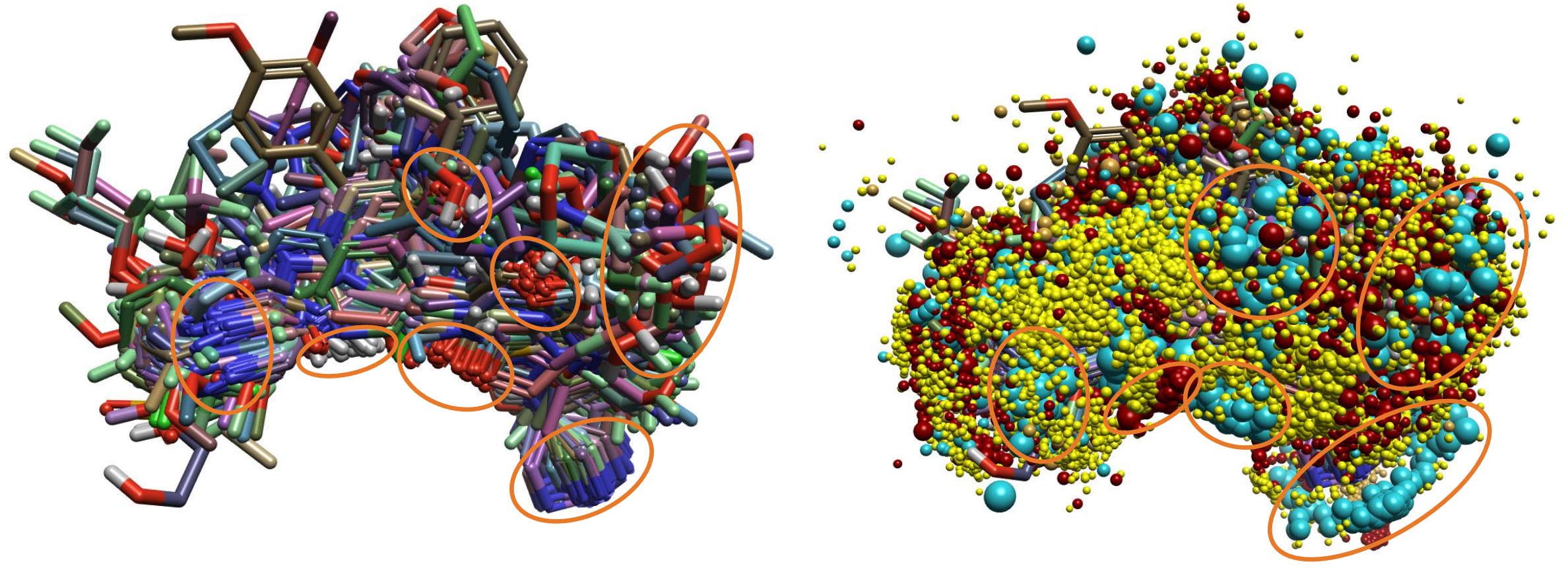
Figure 6. ‘Normal’ alignment of 1159 ligands to the three most active references from each structure subset. (Left) Alignment of 3D structures. (Right) Field points toggled on. Key functional groups (left) and areas of field clusters (right) are highlighted.
Although the alignment of the 1159 ligands looks somewhat disordered, there are several areas where key functional groups are very tightly aligned, such as pyridine/pyrimidine nitrogens, secondary amide nitrogens, amide carbonyls, and hydroxyls/ethers. These alignments become even more visually prominent when field points are toggled, and clusters of positive and negative fields are visible.
Observing ligands individually allows for better visualization of these alignments. Figure 7 shows one ligand from each substructure set superposed to three of the nine references, one from each substructure set, in the active site.
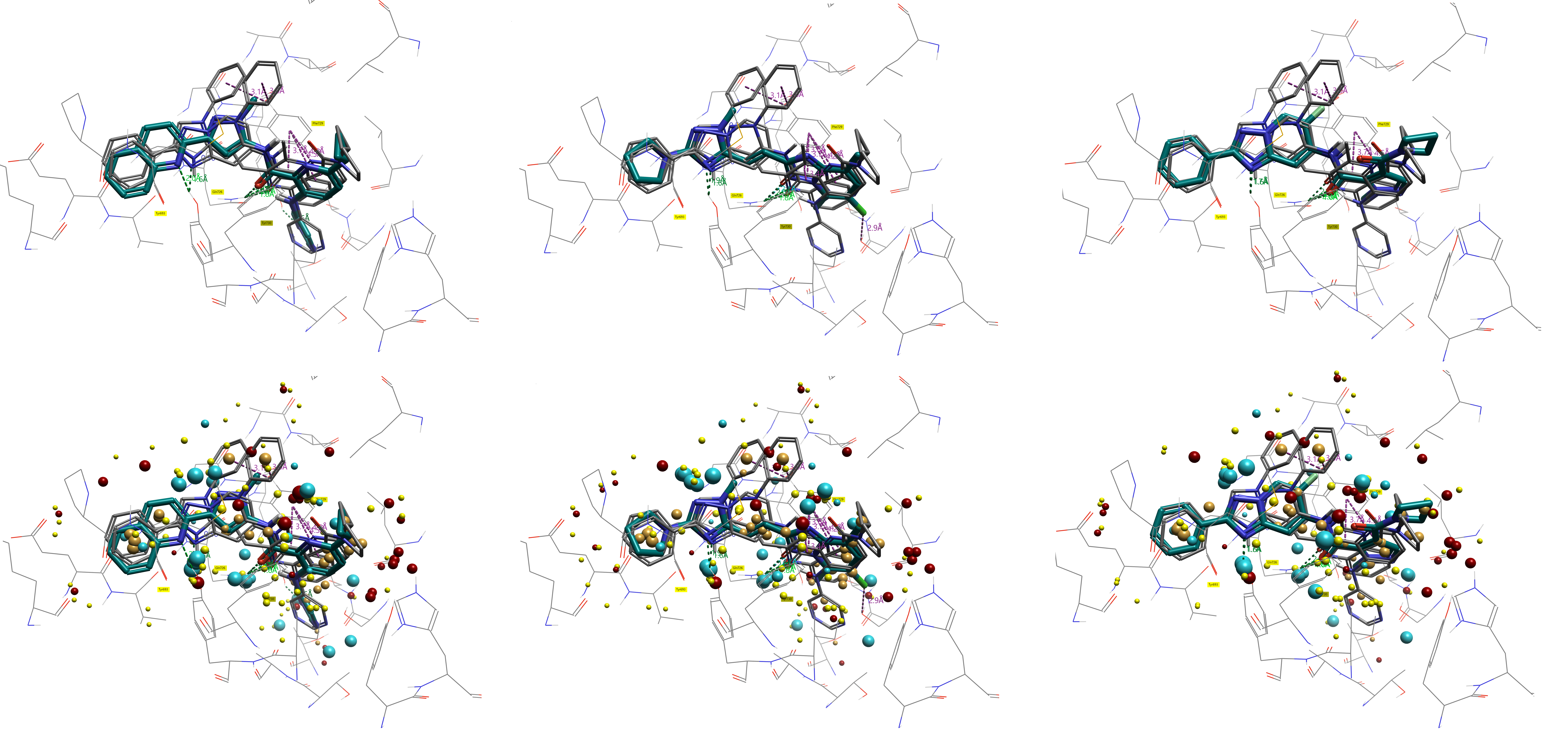 Figure 7. Active site of 5SE7 with aligned ligands (teal) superposed to three references (gray). (Left) aminohetaryl_c1_amide, ligand 46, pIC50 = 10.1 (Middle) c1_hetaryl_alkyl_c2_hetaryl, ligand 300, pIC50 = 9.48 (Right) aryl_c1_amide_c2_hetaryl, ligand 204, pIC50 = 9.11
Figure 7. Active site of 5SE7 with aligned ligands (teal) superposed to three references (gray). (Left) aminohetaryl_c1_amide, ligand 46, pIC50 = 10.1 (Middle) c1_hetaryl_alkyl_c2_hetaryl, ligand 300, pIC50 = 9.48 (Right) aryl_c1_amide_c2_hetaryl, ligand 204, pIC50 = 9.11
From Figure 7, it’s clear that the alignment was successful despite the differences in 2D structure. Not only are the 2D structures relatively well aligned, but the field point clusters are apparent. Furthermore, the key interactions with residues in the active site are retained: all three ligands make hydrogen bonds with Tyr693 and Gln726 and aromatic interactions with Phe729 as do the crystal structure reference ligands.
Structure-based technique: docking ligands for a QSAR study
Because the original study by Tosstorff et al.5 used docked poses with multi-template docking to run the QSAR predictions, this QSAR study was repeated with docked ligands using Flare’s single-template option to seed the docking runs. For each ligand subset, the most active corresponding reference ligand was used to seed the docking results. Ensemble Docking was also performed on the previously mentioned nine crystal structures with the highest RMSD deviations to allow for maximum variation of the protein structure. Figure 8 shows the superimposition of the docked structures for each ligand subset. There was, unsurprisingly, significantly more variation with the docked poses than with the ligand-based alignments, as the ligands had more freedom to complement the protein within the specified docking region (a 6.0 Å active site energy grid was defined by a 6Å radius around the chosen reference ligand).
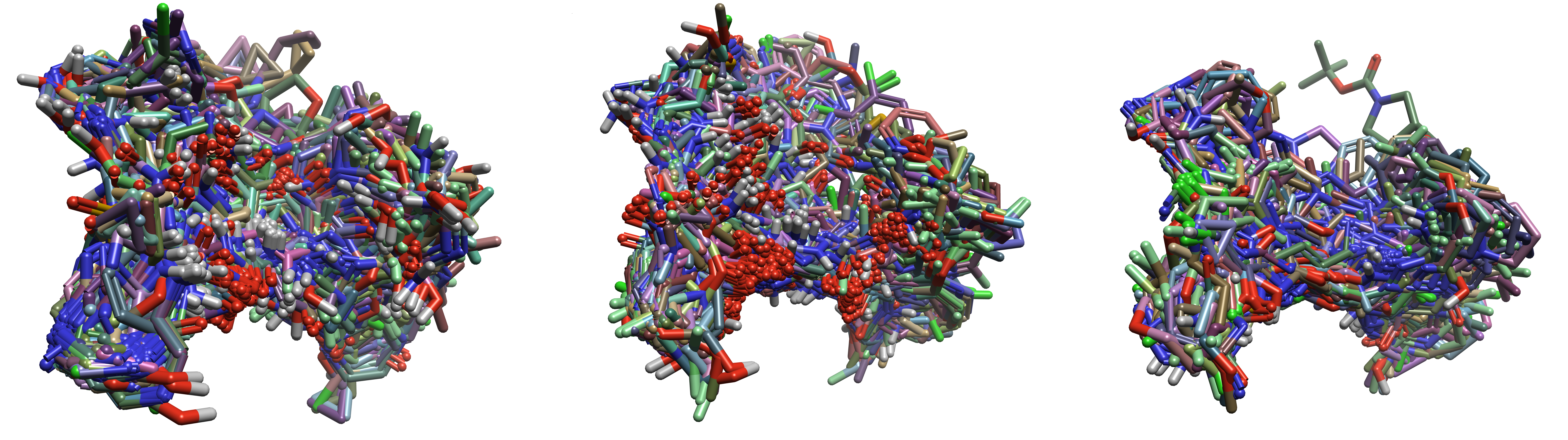
Figure 8. Superimposed docked poses for each ligand subset. (Left) aminohetaryl_c1_amide, (Middle) aryl_c1_amide_c2_hetaryl, (Right) c1_hetaryl_alkyl_c2_hetaryl
QSAR regression model building
Before building QSAR models, 10% of each subset (or 20% of the full dataset for ‘Normal’ ligand alignment’) was removed for a validation set, and the remaining ligands were partitioned into training and test sets by a 70/30 ratio and an even range of activity. The divisions of training/test/validations for MCS ligand alignment and for docking were as follows: aminohetaryl_c1_amide into 285/122/45, aryl_c1_amide_c2_hetaryl into 264/113/42, and c1_hetaryl_alkyl_c2_hetaryl set into 181/78/29. Since the ‘Normal’ ligand alignment did not divide the ligands by structure subset, 20% of the ligands were removed for a validation set, and the rest were split into a training and test set. The final split was 649/278/232 as training/test/validation. Three ligands of the original 1162 were removed due to the SMILES containing two separate ligands as a racemic mixture.
Flare’s Automatic Regression option was used to build all possible regression models, namely Support Vector Machine (SVM), Random Forest (RF), Gaussian Process (GP), k-Nearest Neighbors (kNN), Multilayer Perceptron (MLP), and select the one with the best results. The Consensus model was explored as well, which calculates the average of the predictions from the five individual regression models. An additional model available in Flare is Field QSAR, a regression method that uses Cresset’s 3D descriptors for shape and electrostatics to build not only a predictive model, but also a 3D visualization that illustrates where steric bulk and more positive/negative electrostatics contribute to activity. Field QSAR experiments were also explored in this study. Table 1 shows the QSAR results, including the model giving the best predictive performance and the resultant correlations, as well as the correlation and root mean square error (RMSE) of the validation sets. Field QSAR uses a leave-one-out cross validation method but does not output a separate training set cross-validation correlation.
Results
QSAR for the MCS ligand alignment
The first round of QSAR model building focused on the MCS aligned ligands. The results are in Table 1.
Table 1. QSAR models for each MCS aligned ligand subset.
| Entry | Ligand subset | QSAR model | r2 Training set |
r2 Training set CV |
r2 Test set |
r2 Validation set |
RMSE Validation set |
| 1 | aminohetaryl_c1_amide | Support Vector Machine |
0.94 | 0.55 | 0.64 | 0.48 | 0.88 ± 0.03 |
| 2 | Consensus | 0.91 | 0.53 | 0.60 | 0.48 | 0.88 ± 0.03 | |
| 3 | Field | 0.71 | 0.48 | 0.56 | 0.31 | 1.01 ± 0.04 | |
| 4 | aryl_c1_amide_c2_hetaryl | Support Vector Machine | 0.93 | 0.50 | 0.51 | 0.58 | 0.73 ± 0.03 |
| 5 | Consensus | 0.90 | 0.49 | 0.53 | 0.63 | 0.70 ± 0.02 | |
| 6 | Field | 0.52 | 0.40 | 0.42 | 0.60 | 0.71 ± 0.02 | |
| 7 | c1_hetaryl_alkyl_c2_hetaryl | Support Vector Machine |
0.98 | 0.50 | 0.47 | 0.25 | 0.88 ± 0.03 |
| 8 | Consensus | 0.97 | 0.51 | 0.44 | 0.27 | 0.83 ± 0.03 | |
| 9 | Field | 0.75 | 0.50 | 0.43 | 0.34 |
0.85 ± 0.03 |
From Table 1, it’s clear that despite the high variation in ligand structure alignment, the predictive models performed reasonably well. Although the correlation between predicted and actual activity is not ideal for the c1_hetaryl_alkyl_c2_hetaryl subset, the RMSE, representing the difference between the predicted and actual activities in terms of the dataset size, is quite low for all ligand subsets. On average, the difference between predicted and actual activities calculated by either regression models or Field QSAR is less than 1 pIC50 value (RMSE average = 0.83). This data is very promising for our ligand-based approach to activity prediction and outperforms the structure-based methods tested by Tosstorff et al. (RMSE = 1.04 – 3.31) when evaluating all ligands randomly, across substructure tags, and by temporal splitting.5
To see if the results could be improved, the aminohetaryl_c1_amide dataset was inspected, and the alignments were individually adjusted against the reference ligands by rotating individual bonds. The predictivity of the model only increased by r2 of 0.02, or three percent. This adjustment was repeated for the smallest dataset, the c1_hetaryl_alkyl_c2_hetaryl ligands, but the predictivity increased again by r2 of 0.02, or five percent. This negligible improvement highlights that there is confidence in Flare’s alignment, despite the large visual discrepancy in structural variation within the data subsets.
QSAR for the electrostatic field- and shape-based alignment
The QSAR model results for the ‘Normal’ ligand alignment are in Table 2 and Figure 9.
Table 2. QSAR models for docked ligands
| Entry | QSAR model | r2 Training set |
r2 Training set CV |
r2 Test set |
r2 Validation set |
RMSE Validation set |
| 1 | Support Vector Machine | 1.00 | 0.46 | 0.51 | 0.50 | 0.83 ± 0.01 |
| 2 | Consensus | 0.97 | 0.46 | 0.50 | 0.53 | 0.82 ± 0.01 |
| 3 | Field | 0.61 | 0.33 | 0.40 | 0.46 | 0.87 ± 0.01 |
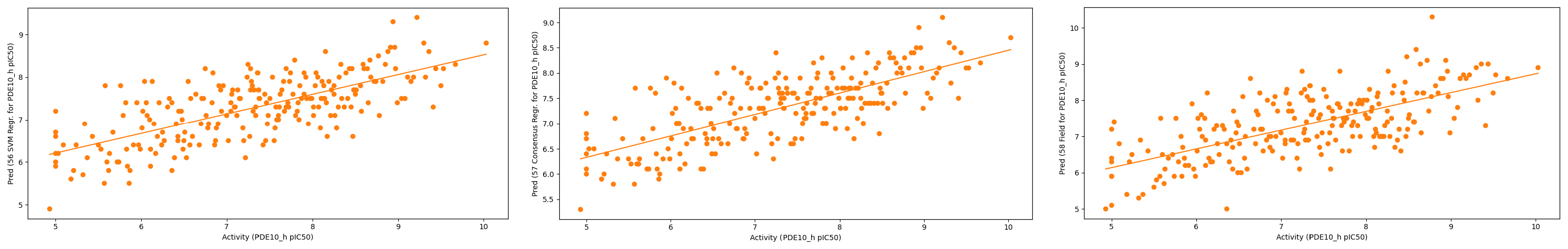
Figure 9. Plots of predicted vs actual activity for the validation set in the ‘Normal’ alignment. (Left) r2 = 0.50 Support Vector Machine; (Middle) r2 = 0.53 Consensus; (Right) r2 = 0.46 Field QSAR.
The QSAR data further supports the notion that ligand alignment provides more robust QSAR models, as exhibited by this diverse dataset. Furthermore, the results from the ‘Normal’ alignment of all 1159 compounds are comparable to those of the MCS alignment of each individual substructure set, demonstrating the value of the Cresset 3D shape and electrostatic descriptors. The average RMSE values of the ‘Normal’ and MCS alignments are comparable: the average RMSE values are 0.83 and 0.84, respectively. For this particular dataset, the activity of new ligands can be reliably predicted within 1 pIC50 value when MCS or ‘Normal’ alignments are performed on new prediction sets.
QSAR for the docked ligands
The QSAR model-building was performed similarly to the ligand-based experiments Table 3 displays the results.
Table 3. QSAR models for docked ligands
| Entry | Ligand subset | QSAR model | r2 Training set |
r2 Training set CV |
r2 Test set |
r2 Validation set |
RMSE Validation set |
| 1 | aminohetaryl_c1_amide | Support Vector Machine |
0.98 | 0.29 | 0.45 | 0.47 | 0.83 ± 0.02 |
| 2 | Consensus | 0.94 | 0.30 | 0.46 | 0.47 | 0.83 ± 0.02 | |
| 3 | Field | 0.69 | 0.24 | 0.37 | 0.27 | 1.02 ± 0.03 | |
| 4 | aryl_c1_amide_c2_hetaryl | Support Vector Machine | 0.85 | 0.23 | 0.30 | 0.34 | 0.98 ± 0.03 |
| 5 | Consensus | 0.89 | 0.22 | 0.30 | 0.34 | 0.98 ± 0.03 | |
| 6 | Field | 0.49 | 0.19 | 0.18 | 0.14 | 1.13 ± 0.03 | |
| 7 | c1_hetaryl_alkyl_c2_hetaryl | Support Vector Machine |
0.96 | 0.36 | 0.37 | 0.31 | 0.96 ± 0.04 |
| 8 | Consensus | 0.92 | 0.34 | 0.35 | 0.28 | 0.97 ± 0.04 | |
| 9 | Field | 0.62 | 0.23 | 0.17 | 0.22 |
1.04 ± 0.03 |
The lower correlations of the test sets for the docking methods are not surprising, given that the superimpositions of the docked ligands are much less ordered than the ligand-based alignments. However, the maximum difference between the structure-based and ligand-based ligand preparation demonstrates an RMSE of 1.13 (aryl_c1_amide_c2_hetaryl), which is still impressive considering the large difference in 3D structural alignment. In general, 3D QSAR models using ligand datasets prepared via ligand-based alignment instead of by molecular docking are confirmed to produce more reliable QSAR models. This conclusion is supported by the results of these QSAR experiments: 3D QSAR models built from ligand-based alignments are more reliable in predicting anti-PDE10A activity than models built from docked ligands. Furthermore, the structure-based docking experiments took considerably longer to run than ligand alignments given the complexity of the experiments.
Conclusion
Activity prediction is a highly sought-after tool in drug discovery, and many ligand-based and structure-based methods exist to aid in these endeavors. When a protein structure is available, many research groups focus on structure-based methods such as free energy perturbation (FEP), molecular dynamics (MD), or molecular docking. When a protein structure is unavailable, ligand-based methods must be used, such as pharmacophore modeling or QSAR. However, in this case, the utilization of both ligand and structural information led to synergistic QSAR results; crystallographic ligand references were used to guide the ligand alignment, resulting in highly reliable QSAR models.
Reliable QSAR models require robust and reliable ligand datasets, preferably from the same research group or institution and tested using the same biochemical assays. Fortunately, the published dataset5 fit these criteria and allowed several QSAR experiments to be conducted. Using Cresset’s ligand-based alignment methods and 3D descriptors for ligand electrostatics and shape, the resultant predictive models can accurately predict ligand activity within 1 pIC50 value, demonstrating the robustness of the methods.
Although there is no one correct way to analyze a dataset and generate models to predict ligand activity, this study has shown that QSAR was consistently reliable for this set of PDE10A ligands.
References
- Flare™, version 7, Cresset®, Litlington, Cambridgeshire, UK; http://www.cresset-group.com/flare/; Cheeseright T., Mackey M., Rose S., Vinter, A.; Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation J. Chem. Inf. Model. 2006, 46 (2), 665-676; Bauer M. R., Mackey M. D.; Electrostatic Complementarity as a Fast and Effective Tool to Optimize Binding and Selectivity of Protein–Ligand Complexes J. Med. Chem. 2019, 62, 6, 3036-3050; Maximilian Kuhn, Stuart Firth-Clark, Paolo Tosco, Antonia S. J. S. Mey, Mark Mackey and Julien Michel Assessment of Binding Affinity via Alchemical Free-Energy Calculations J. Chem. Inf. Model. 2020, 60, 6, 3120–3130
- Meng, X.-Y., et al. Molecular Docking: A powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 2011, 7(2), 146–15.7. doi.org/10.2174/157340911795677602
- Cheeseright, T., et al. Molecular Field Extrema as Descriptors of Biological Activity: Definition and Validation. J Chem Inf Model 2006, 46, 2, 665–676. doi.org/10.1021/ci050357s
- Verhoest, P. R., Chapin, D. S., Corman, M. et al. Discovery of a novel class of phosphodiesterase 10A inhibitors and identification of clinical candidate 2-[4-(1-methyl-4-pyridin-4-yl-1H-pyrazol-3-yl)-phenoxymethyl]-quinoline (PF-2545920) for the treatment of schizophrenia. J Med Chem 2009, 52, 16, 5188–96. doi.org/10.1021/jm900521k
- Tosstorff, A., Rudolph, M. G., et al. A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios. J Comp-Aided Molec Des 2022, 36, 753–765.