Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
As those of you who have been regularly reading the Cresset newsletter will know, we have had an ongoing collaboration with Simon Mcintosh-Smith at the University of Bristol to bring some of his group’s expertise in parallel and multicore computing to Cresset. The initial aim of the collaboration is to port our field based alignment and similarity algorithm to run on graphics cards (GPUs). This was no small task as it involved rewriting a number of highly advanced and complicated algorithms into openCL, a new language designed explicitly to allow massively parallel computation.
GPUs are strange beasts. The architecture was originally designed for shunting vast numbers of triangles and textures around in order to make computer games prettier. They have increasingly become useful for more general-purpose computing tasks, but their origins mean that while they can be blazingly fast at some types of computation they do other things very poorly. Provided you can tweak your algorithm into an appropriate form you can get remarkable performance out of a single cheap graphics card, but not all algorithms are amenable to such tweaking and it takes an expert to get the best results. Luckily we have an expert on hand in the form of Simon Krige, who joined us last year from Bristol!
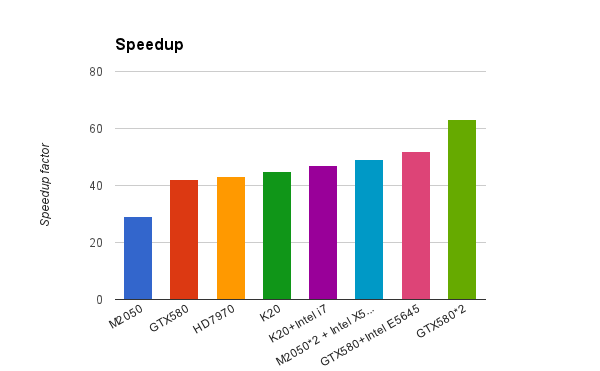
Simon has been working hard rewriting the core Blaze functionality into openCL, from our initial field point clique-matching algorithm to shape similarity calculations to the core field similarity computations that are at the center of Cresset’s ability to sensibly compare molecules’ electrostatic properties. The results look set to dramatically change the landscape for field based virtual screening. As you can see from the graph below, a single NVidia or AMD graphics card has the same screening performance as more than 40 modern CPU cores!

BlazeGPU is scheduled for release later this year. Contact us for more information on Blaze or BlazeGPU.