The adaptability of pyFlare to access advanced data visualization and calculation functionalities
We showcase some examples within a drug discovery context where the pyFlare environment has been further expanded with advanced data ...
News
Docking in Flare™ is one of the platform’s most popular functions, providing its users with detailed feedback on new molecule designs, high enrichment in virtual screening, and excellent pose prediction. Until recently, the docking experiments that were available were ‘Normal’, ‘Covalent’, and ‘Ensemble’. Normal docking is the standard docking feature: a ligand docked to a protein within a specified spatial energy grid. Covalent docking adds the formation of a covalent bond between a nucleophilic residue of the protein and a reactive warhead in the ligand. Finally, Ensemble docking allows the user to apply Normal docking to several protein conformations at once.
A more recently added feature is ‘Ensemble Covalent’ docking, the option that combines Covalent and Ensemble: this new feature allows the user to apply covalent docking to several proteins at once.
Before starting an Ensemble Covalent docking experiment, it is necessary to first prepare, sequence align and superpose your proteins. Protein preparation can be conveniently performed when importing the protein structures, which can then be sequence aligned using the ‘Align’ function under the ‘Sequences’ tab (Figure 1). The ‘Superpose’ function will then superpose the proteins in 3D so that they can be used in Ensemble Covalent docking:
 Figure 1: Selecting the ‘Superpose’ function within Flare.
Figure 1: Selecting the ‘Superpose’ function within Flare.
Once the proteins are prepared, aligned and superimposed in 3D, select all the proteins in the ‘Proteins’ table to which you would like to dock your ligands. The Ensemble Covalent docking experiment can be found under the ‘3D Pose’ tab and ‘Dock’ dropdown menu:
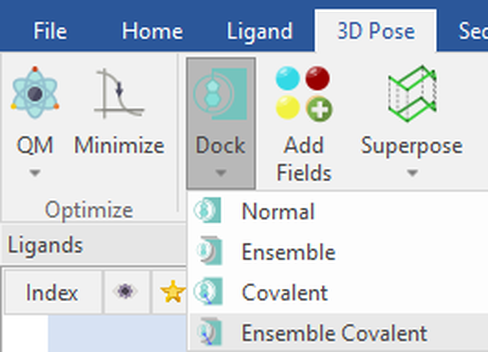 Figure 2: Ensemble Covalent docking workflow selected from the ‘Dock’ dropdown menu in Flare.
Figure 2: Ensemble Covalent docking workflow selected from the ‘Dock’ dropdown menu in Flare.
In the Ensemble Covalent Docking calculation panel (Figure 3), you can then easily set the region where to dock your ligands by using the position of a crystallographic ligand or by picking atoms from the active site, as well as choose a representative catalytic nucleophilic residue for the proteins. Here, we’ve chosen Ser554 of a enzyme prolyl oligopeptidase, and Flare will recognize the equivalent catalytic serine residues for the other aligned proteins by 3D proximity. Alternatively, when aligning proteins that have different catalytic residues, you may select the covalent residue in each protein with the Residue dropdown menus:
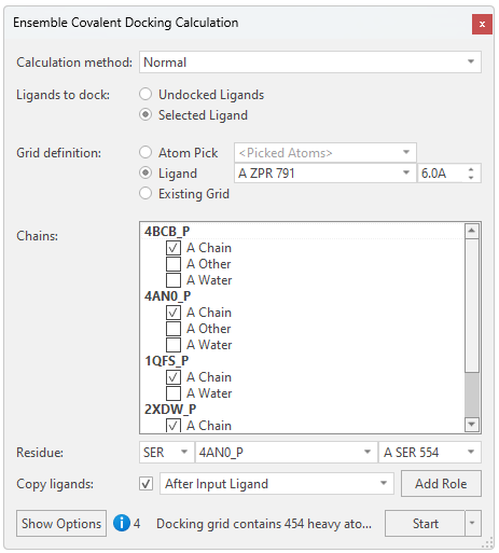
Figure 3: Setting up the Ensemble Covalent docking experiment in Flare.
The Ensemble Covalent docking experiment is now ready to start!
To illustrate this new Flare feature, we will use compound KYP-2047, a prolyl oligopeptidase (POP, or PREP) inhibitor which is currently being utilized to study the role of POP in neurodegenerative diseases,1 macular degeneration,2 and glioblastoma.3 Figure 4 shows the 2D and 3D structure of this molecule.
KYP-2047 has been co-crystallized with POP,4 and a structure is available from the RCSB as pdb: 4AN0. Figure 4 (Right) shows the protein surface and interacting residues in the active site of POP, with serine 554 covalently bound to KYP-2047.

Figure 4. (Left) 2D structure of KYP-2047 with associated POP5 activity: the covalent warhead is highlighted. (Right) KYP-2047 covalently bound to POP (pdb 4AN0). Protein-ligand interactions are shown with default Flare coloring: green = strong hydrogen bond, gray = hydrophobic contacts. The protein surface is colored by hydrophobicity6: blue = hydrophilic, beige = hydrophobic.
One way to utilize the Ensemble Covalent docking feature is to mimic ‘flexible’ docking into one protein. One of the possible ways to do this is to minimize or modify the 3D conformation of individual residues and observe the effect on the docking poses: changing one residue at a time allows for a more controlled experiment. In the case of this protease, one of the key residues for substrate and inhibitor stabilization in the active site is Arg643. Here, we have slightly rotated the side chain to allow for variation in the conformation of the active site (Figure 5, left). KYP-2047 was then docked to five different conformations of POP, varying only in position of the arginine side chain (Figure 5, right). The top-scoring poses are listed in the Ligands table, ranked by LeadFinder™ Rank Score. The associated proteins are listed in the Protein column (Figure 6).
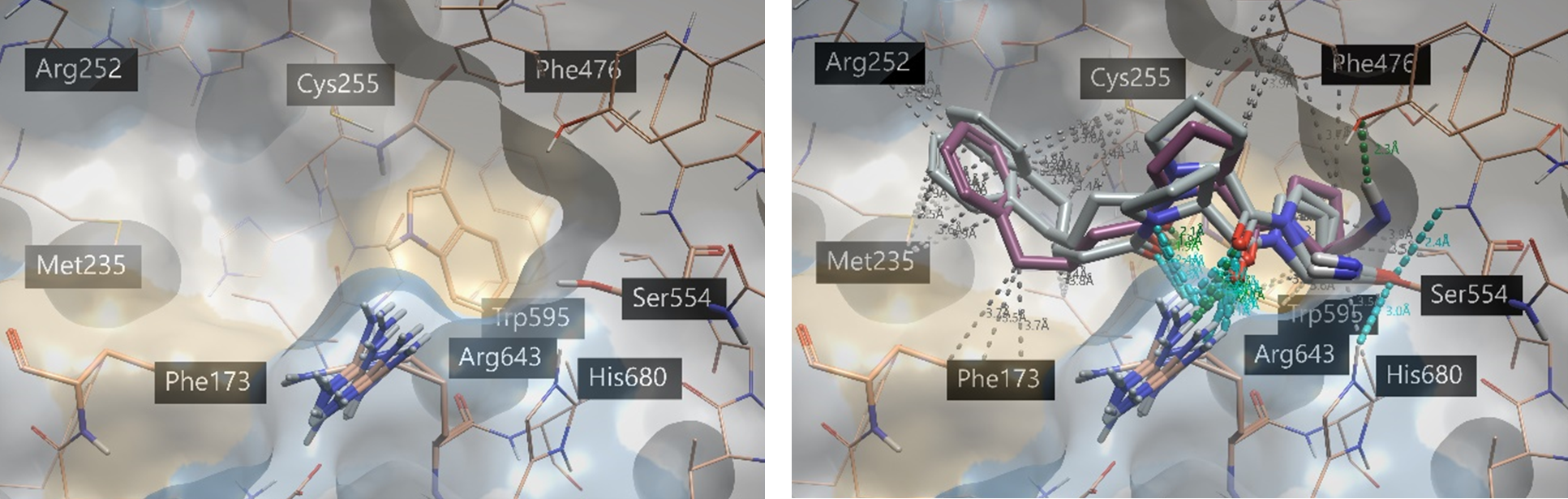
Figure 5. (Left) Active site of POP (pdb 4AN0), including arginine rotated in several conformations. (Right) Top two poses of KYP-2047 (gray) docked to all conformations with Ensemble Covalent docking. The co-crystallized ligand is shown in violet. Protein-ligand interactions are shown with default Flare coloring: green = strong hydrogen bond, blue = weak hydrogen bond, gray = hydrophobic contacts. The protein surface displays hydrophobicity6: blue = hydrophilic, beige = hydrophobic.
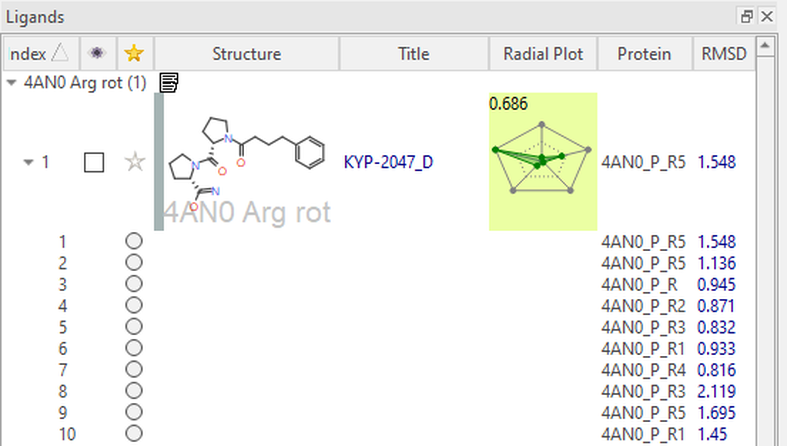
Figure 6: Ensemble covalent docking experiment results displayed within the proteins column in Flare.
In Figure 5, We can see that the top two docked poses of KYP-2047 consistently align with the co-crystalized ligand and make the necessary key interactions in the active site (RMSD = 1.5 Å, 1.1 Å). The 1,4-dicarbonyl substructure hydrogen bonds with Trp595 and Arg643, despite the varying conformations of the latter residue. We can also see that there are many hydrophobic contacts made with the largely hydrophobic binding pocket. As well as the example shown here, it is also possible to generate different protein structures where the side-chain conformation of the nucleophilic residue is explored.
Another way to approach the ‘flexible’ docking method is to simply dock to several X-ray structures of the same protein at once. Every 3D model is slightly different, giving some variation in the 3D structure of the active site. For example, there are many structures of POP in the Protein Data Bank. Upon aligning just four of these structures (Figure 7, 4AN0, 2XDW, 1QFS, 4BCB), we can see that there is slight variation in 3D coordinates of key active site residues.
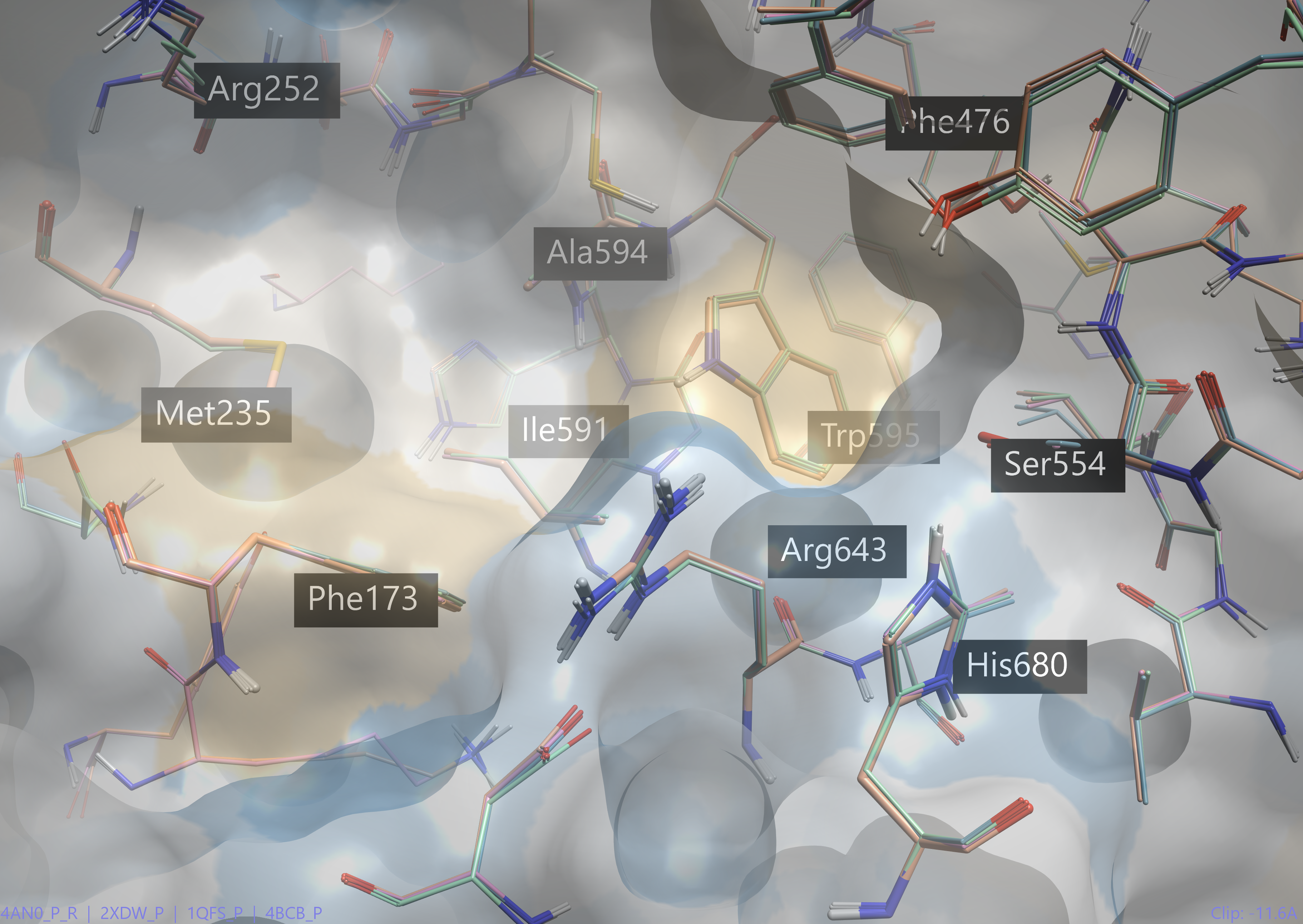 Figure 7. Key active site residues of four crystal structures of POP (pdb 4AN0, 2XDW, 1QFS, 4BCB).
Figure 7. Key active site residues of four crystal structures of POP (pdb 4AN0, 2XDW, 1QFS, 4BCB).
Running Ensemble Covalent docking of KYP-2047 on these prepared proteins yields KYP-2047 docked to all the crystal structures. Figure 8 shows the 3D View of the top pose compared to the co-crystallized KYP-2047.
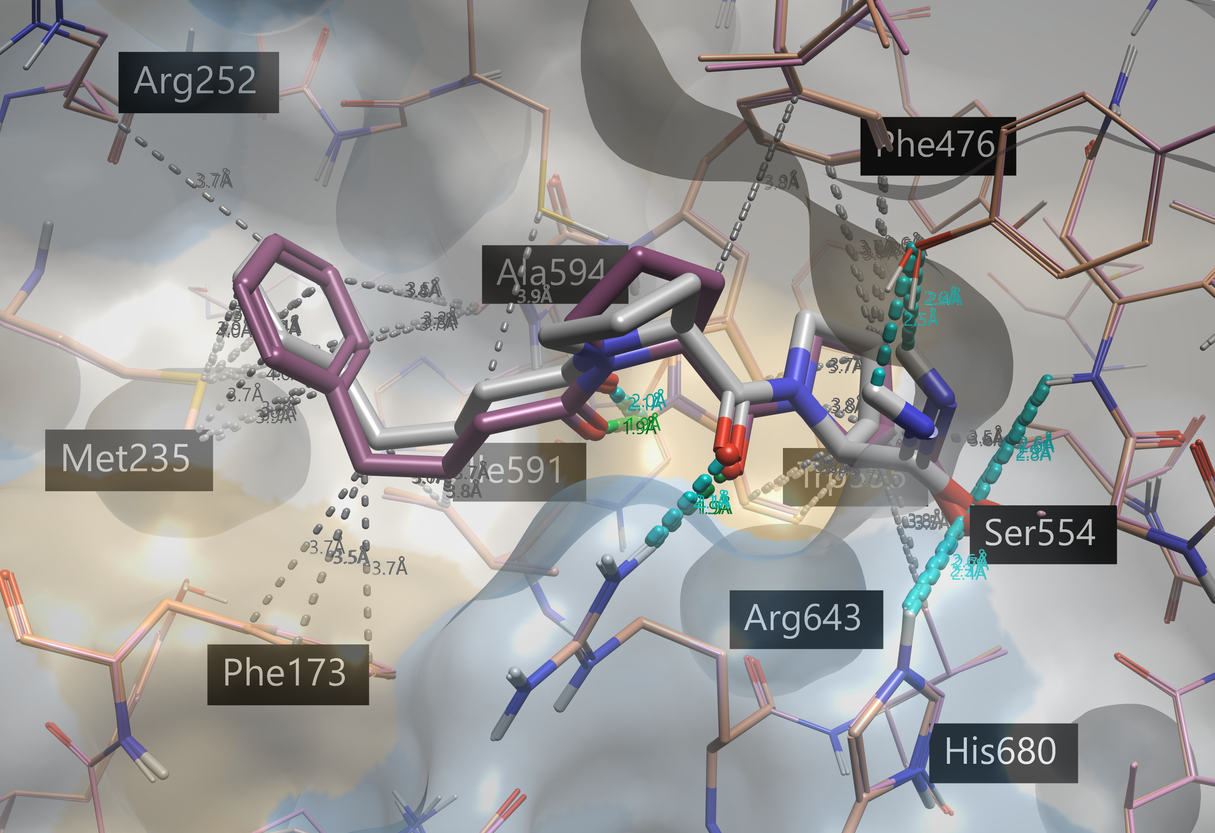 Figure 8. Top-scoring pose of docked KYP-2047 (gray), using Ensemble Covalent docking on four POP structures (associated pdb 4BCB is shown in pink). The co-crystallized KYP-2047 (associated pdb 4AN0, orange) is shown in violet. Protein-ligand interactions are shown with default Flare coloring: green = strong hydrogen bond, blue = weak hydrogen bond, gray = hydrophobic contacts. The protein surface displays hydrophobicity: blue = hydrophilic, beige = hydrophobic.
Figure 8. Top-scoring pose of docked KYP-2047 (gray), using Ensemble Covalent docking on four POP structures (associated pdb 4BCB is shown in pink). The co-crystallized KYP-2047 (associated pdb 4AN0, orange) is shown in violet. Protein-ligand interactions are shown with default Flare coloring: green = strong hydrogen bond, blue = weak hydrogen bond, gray = hydrophobic contacts. The protein surface displays hydrophobicity: blue = hydrophilic, beige = hydrophobic.
The top six pose results (with at least one pose being generated in each of the protein structures) were very similar to the co-crystallized ligand (RMSDs < 1.5 Å). Like the co-crystallized KYP-2047, the 1,4-dicarbonyl is forming hydrogen bonds with the tryptophan and arginine residues, while the benzyl group participates in hydrophobic interactions and aromatic interactions.
In either case above, we can use Ensemble Covalent docking to dock our ligand(s) of interest to each protein of interest and ask Flare to output the best poses for each protein (as opposed to best overall poses). To access this option, click ‘Show Options’ in the Ensemble Covalent Docking Calculation window:
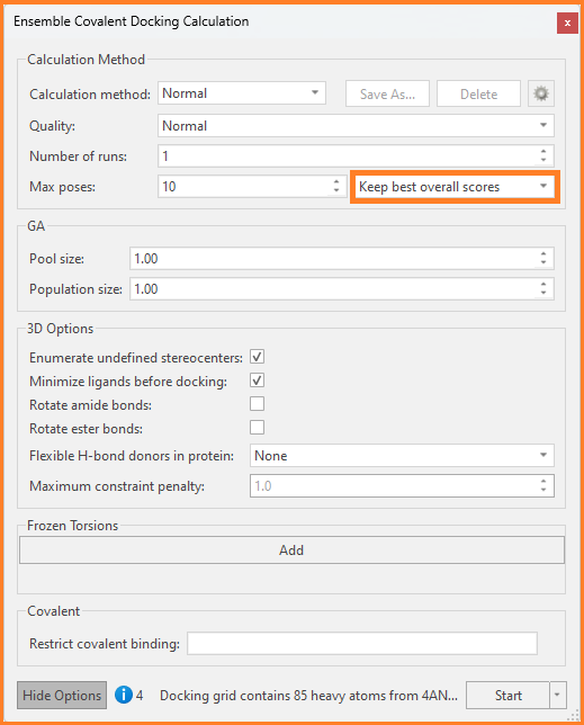 Figure 9: The Ensemble Covalent Docking Calculation window in Flare.
Figure 9: The Ensemble Covalent Docking Calculation window in Flare.
As illustrated with KYP-2047, Ensemble Covalent docking in Flare can be utilized either by docking to several different conformations of one protein or to different pdbs of the same protein.
If you’d like to test the Ensemble Covalent docking feature on your project, while having access to the full portfolio of molecular modeling features within Flare, you can request a free evaluation today. As part of the evaluation process, you’ll receive full support in installing the platform and accessing its wide range of features, while having the freedom to publish any results produced and use these for further research. (If you have any technical questions during your own Ensemble Covalent docking experiment, please do not hesitate to reach out to Cresset Support.)