Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Spark is a bioisostere replacement solution that enables drug discovery research chemists to generate highly innovative ideas to explore chemical space and escape IP and toxicity traps. The problem it solves is quite simple to explain: “This molecule is great! I love it! However, I don’t like that bit.” There can be many reasons why you need to replace part of a molecule: poor physicochemical or ADMET properties, stability, off-target selectivity, patent problems, or even just that you need a backup series. In all of these cases, you’re looking for a change in chemical structure, but ideally without any significant detrimental effect on activity. Spark solves this problem by finding alternative chemistries for you that preserve the properties you want (shape, electrostatics, and hopefully affinity) while changing the things that need changing (chemistry and patentability).
 The Spark process is, at its core, a search for bioisosteres. The exact definition of a bioisostere is rather fuzzy, but the Wikipedia entry is as good as any: 'bioisosteres are chemical substituents or groups with similar physical or chemical properties which produce broadly similar biological properties to another chemical compound.'. Finding bioisosteres ought to be easy - look at the piece you want to replace, search a database for something largely the same size and geometry, and present the results. Simple, right? However, if you delve into it, the whole process is much more complex than it appears.
The Spark process is, at its core, a search for bioisosteres. The exact definition of a bioisostere is rather fuzzy, but the Wikipedia entry is as good as any: 'bioisosteres are chemical substituents or groups with similar physical or chemical properties which produce broadly similar biological properties to another chemical compound.'. Finding bioisosteres ought to be easy - look at the piece you want to replace, search a database for something largely the same size and geometry, and present the results. Simple, right? However, if you delve into it, the whole process is much more complex than it appears.
The first question is how to approach scoring a new candidate fragment. If I’m looking for a replacement for a triazolothiazole, then when I’m presented with a candidate fragment the obvious thing to do is ask “How similar is it to triazolothiazole?”. This approach is flawed. The reason is that the properties of the candidate fragment (and the initial triazolothiazole) depend on their environment i.e., the rest of the molecule. This is especially true if you are interested in molecular electrostatics, as the electrostatic potential of a molecule is a global property that cannot generally be piecewise decomposed. That is why Spark performs all scoring in product space, not in fragment space (Figure 1).
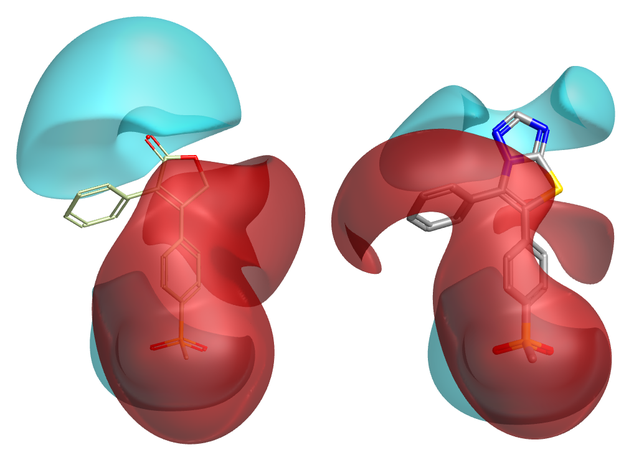
Figure 1: Merging a new fragment can (subtly) change the electrostatics of the rest of the molecule: look at the phenyl on the left.
However, even if you are using a more crude method such as shape or pharmacophore similarity, treating the fragment in isolation can be very misleading, as it may have a steric clash with the rest of the molecule that drastically changes the molecule’s conformation (Figure 2).

Figure 2: Toluyl is similar to phenyl, except when it causes a steric clash. Whether it does or not depends on what R is.
We also need to solve a variety of other cheminformatics and chemistry problems when stitching molecules together:
Having solved all of these problems, the final stage of a Spark search is to score the new merged molecule: how good is it? The standard Spark algorithm uses Cresset’s tried and tested electrostatic and shape similarity algorithm, which gives an excellent assessment of whether the new fragment is indeed bioisosteric to the section it replaced. The power of this scoring algorithm can be attested to by the hundreds of Spark users worldwide.
However, there is one use case that the existing Spark workflow doesn’t fully address. Sometimes, rather than (or in addition to) replacing a section of the target molecule, we instead want to grow it into a hitherto-unexplored section of the binding pocket or make a new interaction with the protein. If there are known ligands that bind into the new pocket then Spark can use these to guide the growth process, but what if there are not? In that case, Spark’s excellent performance in ligand-guided bioisosteric assessment is no use as we don’t have a ligand to guide us.
The solution, introduced in Spark V10.6, is to use docking as a part of the scoring process. The docking score is determined directly from the protein, so there is no longer a need for a reference ligand to inform us of what a good binder in a particular part of the active site should look like. And luckily for us, the Lead Finder™ docking engine has an excellent scoring algorithm with demonstrated ability to find good ligands.
The main issue that we needed to deal with was speed. Spark’s pathway for assessing fragments using ligand similarity is highly optimized and uses a number of well-tested shortcuts to enable us to search large numbers of fragments very quickly. Scoring using the docking engine is inherently significantly slower. Initial testing also showed some potential pitfalls in the docking pathway. Running a free dock of each potential product molecule is just too slow: a typical Spark experiment might look at hundreds of thousands of fragments, and that’s just not possible on a laptop. So, we’re going to have to rely (as the ligand-based scoring pathway does) on keeping the core of the target molecule in place and stitching in a pre-computed set of conformers for the new fragment. The obvious way to proceed was just to use this algorithm but replace the ligand similarity score with a score obtained from the docking scoring function. However, we found that didn’t work very well. The reason is that Lead Finder, like most docking scoring functions, has quite a hard treatment of vdW interactions. Just stitching in the set of fragment conformations and scoring often led to good fragments being discarded because minor vdW clashes led to poor docking scores. Allowing the docker to optimize each pose in the active site fixed this problem, but was again much too slow – you might have 10K fragments, but each has 30 conformers, and each of those might need to be tried with 20 rotamers around a newly-formed single bond: doing a score optimization on each of these possibilities was just not feasible.
So, we need to allow the docker to optimize the pose (even if only slightly), but we can’t afford to do that for all of the conformer/rotamer possibilities for each fragment. How do we do that? The solution came from resurrecting a short-lived research project from some years ago, where we looked at constructing a docking scoring function based on the XED force field and Cresset’s field technology. In essence, we build a pseudo-ligand from the properties of the protein active site, and then use our existing field/shape similarity technology to align molecules to that pseudo-ligand. When we originally looked at this, it performed pretty well, but wasn’t quite up to the standard of the top-performing docking engines at the time. It wasn’t immediately clear how to bridge that gap, so the project was shelved.
However, this was exactly what was needed for bringing docking into Spark - a way of very rapidly selecting the best conformer/rotamer for each candidate fragment, in a way that was forgiving of small steric clashes, was very fast, but gave reasonable results. So, the final algorithm for Spark ended up being a two-stage affair. We first construct our docking 'pseudo-ligand' from the active site and use the existing very rapid Spark algorithm to triage the set of rotamers and conformers for each fragment to the single one that best matches the active site. Then, that final set of structures (one for each candidate fragment) can be scored using Lead Finder, with the Lead Finder genetic algorithm allowed to tweak the molecule slightly to optimize its fit to the active site. The final workflow is still slower than a traditional ligand-based Spark experiment, but much, much faster than doing a free dock of all of the candidate product molecules: you can very definitely do a Spark docking search on databases of hundreds of thousands of fragments on a laptop overnight.
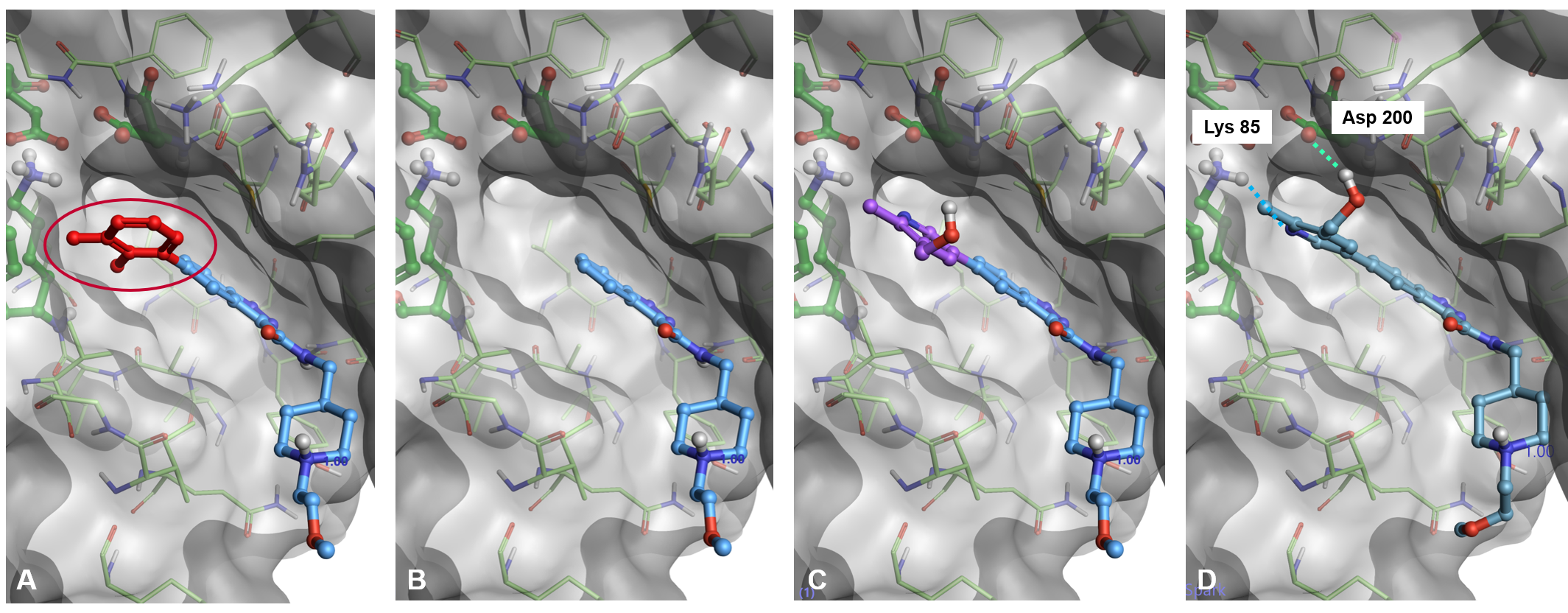
Figure 3: The docking workflow in Spark (PDB: 6TCU). A) A portion of the starter molecule is selected for replacement. B) The selected fragment is deleted. C) A new fragment is attached in a sensible orientation to the truncated starter molecule. D) The pose of the new molecule is optimized using Lead Finder.
For most people, most of the time, the traditional ligand-based Spark experiment is still the way to go, and it will work just as well as it has always done. However, for those times when you don’t have a guiding reference molecule or it’s just not suitable, and you really want to get some suggestions on how to grow into that side pocket that you’ve not been able to access before, then the new Spark docking mode is exactly what you need.
If you would like to try this on your project, request an evaluation and let us know how well it works for you.