Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Hit Expander is a recently introduced component of the molecular modeling platform Flare™, that enables chemists to quickly explore chemical space by manipulating promising molecules with conservative changes. Through the addition of small functional groups, or by performing aromatic single atom substitutions (for example, the conversion of aromatic carbon atoms to aromatic nitrogen), a library of analogues is quickly generated, which can be subsequently analyzed using other Flare components.
Earlier this year, our Application Scientist, Boyli Ghosh led an exclusive online webinar session, during which she presented a case study using Hit Expander to design a new library of analogues, and subsequently predict the relative binding affinity of each against a target protein using predictive QSAR models. This case study provides an example workflow for the facilitation of hit and lead optimization in drug discovery projects.
For those unable to attend the webinar, or interested in catching up, this article serves to summarize the key topics, and highlight the areas of Flare functionality which were demonstrated. We have annotated the headings with the timings of each section within the webinar, for which the recording can be requested, via this link.
Computational methods continue to be used increasingly within drug design to aid with the discovery of new drug candidates. To efficiently combine in silico synthesis with ligand-based drug design, Cresset developed and introduced the Hit Expander module within Flare.
In the case study explored in this webinar, we focused on designing inhibitors of Bruton’s tyrosine kinase (BTK), which is involved in the regulation of B-cell development, activation, and survival through B-cell antigen receptor (BCR) signaling. The inhibition of BTK has the potential to provide therapy for lymphoma, inflammatory disorders, and other autoimmune diseases.
Hit Expander (Figure 1) has been designed to enable the rapid chemical exploration of a single hit compound and works by adding a choice of substituents to all available positions on the selected compound. Aromatic (single atom) substitutions are also performed wherever appropriate.
The ligand variants which are generated through Hit Expander are then available for further experiments within Flare: the most promising molecules can be prioritized with methods such as Free Energy Perturbation (FEP), Electrostatic Complementarity™ (EC) and predictive QSAR models.
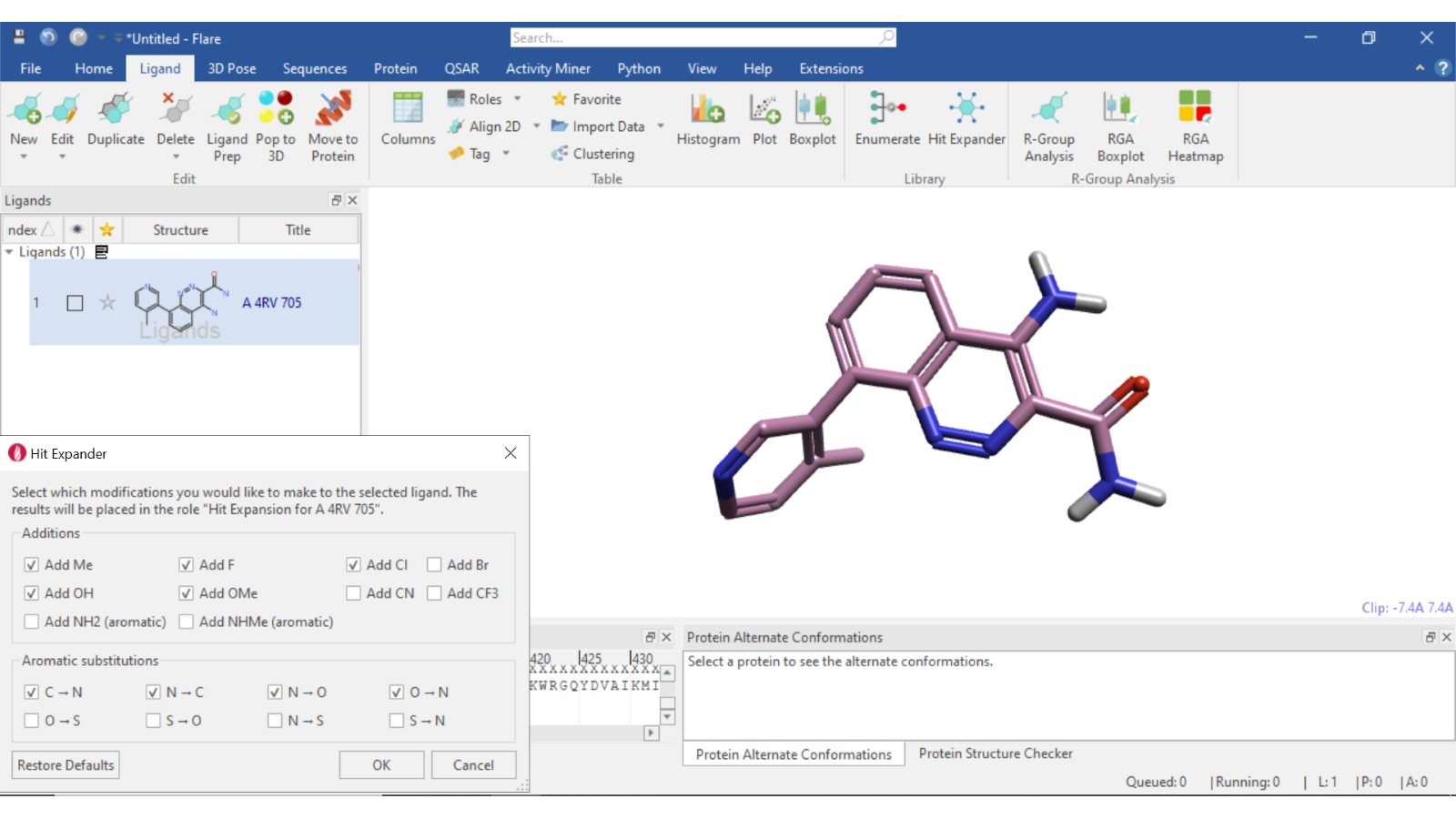
Figure 1: Setting up a Hit Expander experiment within Flare.
For the case study outlined during the webinar, a collection of 90 analogs of BTK inhibitors were selected, originating from US patent 2015/0038510. Figure 2 summarizes the workflow in Flare which was applied to these compounds.
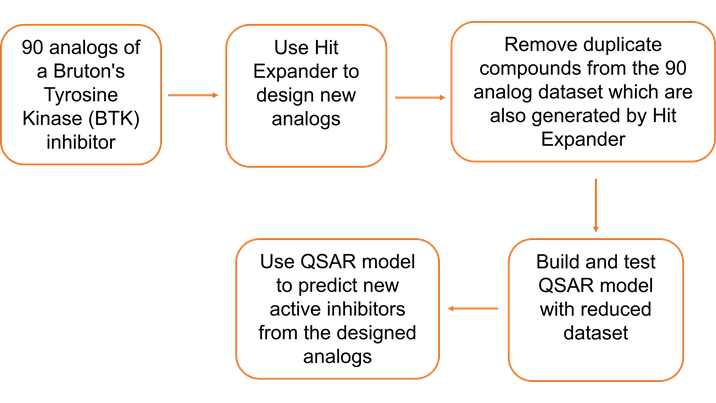
Figure 2: The Flare Hit Expander/ QSAR workflow applied to 90 analogs of a Bruton’s Tyrosine Kinase (BTK) inhibitor.
Cresset’s unique eXtended Electron Distribution (XED) force field differentiates from other traditional force fields, in that it applies charges both to the atom center and to additional monopole ‘XED’ points. This more complex internal electrostatic model means that the force field allows for excellent modeling of polarizing effects, substituent effects, atomic charge anisotropy and aromatic-aromatic interactions.
During this section of the webinar, Boyli provided a background overview of the XED force field, and how it can support enabling a greater understanding in molecular design, through the generation and visualization of field points.

Figure 3: Cresset’s XED force field, applied to show a positive and negative molecular fields.
To produce the QSAR models within Flare, a series of key steps are undertaken, beginning by defining the bioactive conformation of the reference ligands, from known BTK crystal structures. As in this case, this information may be available from existing experimental data, otherwise pharmacophore modeling using FieldTemplater™ in Flare can be used to generate a bioactive conformation hypothesis for your ligand series.
The data set is then aligned to this reference ligand, and visual inspection allows for the identification of molecules which are poorly aligned. Manual intervention is then carried out to ensure that all molecules within the training set are well aligned and ready for the QSAR experiment.
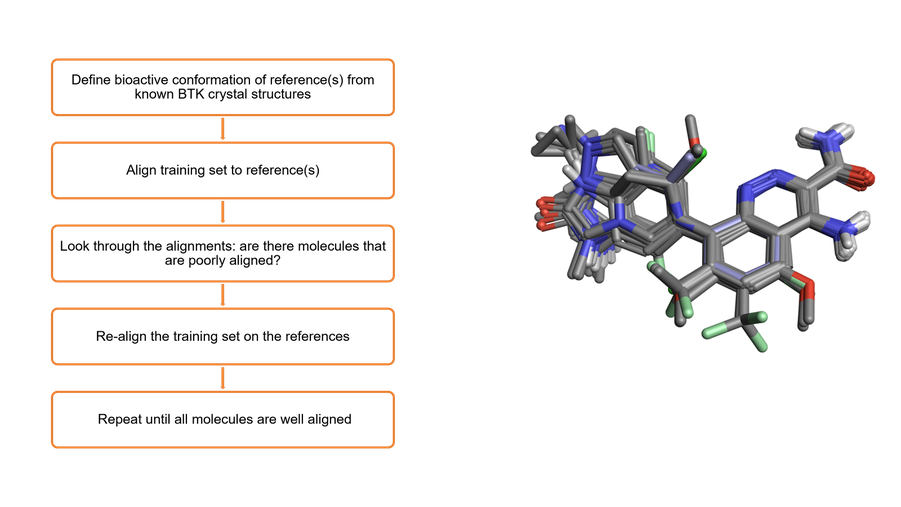
Figure 4: The workflow for 3D alignment of the BTK ligand dataset.
First, using the PDB ID, the target protein is imported into the Flare GUI, and an appropriate protonation state is applied. Flare’s protein preparation process is then initiated, to optimize the protein for the subsequent experiments. The reference ligand is also imported within its crystallographic structure and visualized within the active site of the target protein.
The collection of 90 BTK inhibitors are imported and aligned against the initial reference ligand. These are subsequently portioned into a training set, and test set (30%).
For the Hit Expander experiment, the co-crystallized ligand from the crystal structure (which was included within the test set) was selected as the initial starting point. For this experiment, 4 chosen functional groups (Me, F, Cl and Ome) were selected as the desired modifications. 3 further ligands, one containing an unsubstituted indazole, and others based on their varying activity levels were also selected, to help widen the diversity of hits generated.
The Hit Expansion experiment generated a total of 72 new hits, based on these input designs. Any duplicate designs which existed within the initial portfolio of 90 molecules were removed, to avoid any bias within the QSAR modeling.
The remaining molecules within the training set and test set were selected for a Regression Models QSAR model type. Once the desired settings are applied, the calculation process begins, first by adding fields to the molecular data set.
The remaining molecules within the training set and test set were selected for a Regression Models QSAR model type. Once the desired settings are applied, the calculation process begins, first by adding fields to the molecular data set.
Figure 5: Setting up the QSAR Model experiment within Flare.
The results from the experiment calculated an R2 value of 0.98 for the training set, 0.76 for the cross-validated training set, and 0.66 for the test set. Together, these suggest that the QSAR model has a good predictive activity, and as such can be applied to predict the activity of the compounds generated via Hit Expander.
All the Hit Expander generated compounds were selected and visualized together, which showed a tight clustering of some common functional groups, and key positive and negative electrostatic fields.
The 19 ‘duplicate’ compounds which existed both within the initial dataset of 90 ligands and the generated collection of Hit Expander molecules were selected as favorites. This allowed them to be plotted, displaying their predicted activity against their known activity levels.
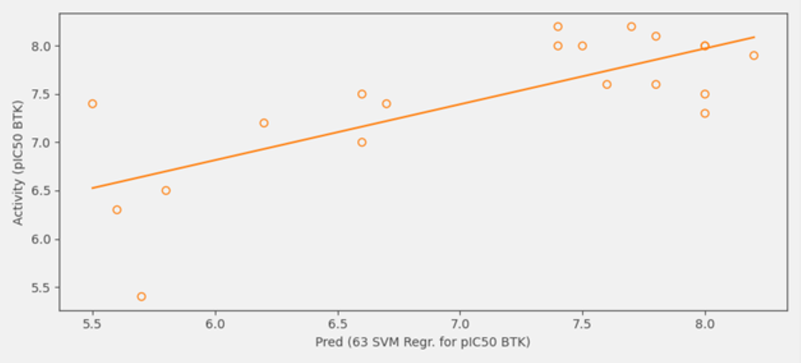
Figure 6: Predicted vs experimental pIC50 values for the 19 ‘duplicate’ compounds.
The R2 value of 0.562 indicates a good correlation between known and predicted activity. With the predictive QSAR model in hand, several new potential active analogs have been identified based on the predicted pIC50 values from the enumerated dataset by Hit expander.
The case study demonstrated that the XED Force Field and QSAR modeling in Flare can be reliably used to build a predictive model for the BTK-inhibitor dataset. Hit Expander helped generate new ideas that were predicted to be nanomolar active by the validated QSAR model.
See the full Flare workflow demonstration, by requesting access to the webinar recording. In doing so, you’ll also receive access to all our recent webinars, covering a full range of computational chemistry techniques.
Try this workflow on your project- request an evaluation of Flare and see how the combined Hit Expander/ QSAR approach can be applied to other ligand-based drug design projects, within which the protein structural information is unavailable.