Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
To obtain accurate and reliable thermodynamic properties of small molecules in Molecular Dynamics (MD) or Free Energy Perturbation (FEP) calculations, it is desirable to improve the molecular description of your molecule beyond the default force field parameterization. In this article, we will introduce how, in Flare™, it is possible to use a hybrid of quantum mechanical (QM) methods: Density Functional Theory (DFT) and an extended semiempirical tight-binding model (GFN2-xTB) in a workflow to calculate custom forcefield parameters for your ligands of interest.
FEP and MD calculations require well-parametrized force fields to describe the bonding and non-bonding potentials of the system. When performing such calculations in Flare, the Open Force Field (OpenFF) is used to describe these potentials; however, what happens when we are interested in modeling a novel molecule not included or poorly represented in the training set on which OpenFF was parameterized? In these cases, OpenFF has reduced transferability due to the limited number of available torsional potential energy parameters, which will inhibit the accessibility of key small molecule phase space that could be critical for protein binding. Torsion parameters are crucial for realistically describing the small molecule’s conformational distribution.
In Flare, it is possible to generate custom force field parameters for all torsions of the molecules under investigation using a combination of ab initio (DFT) and semiempirical quantum mechanical (GFN2-xTB) methods (Figure 1).
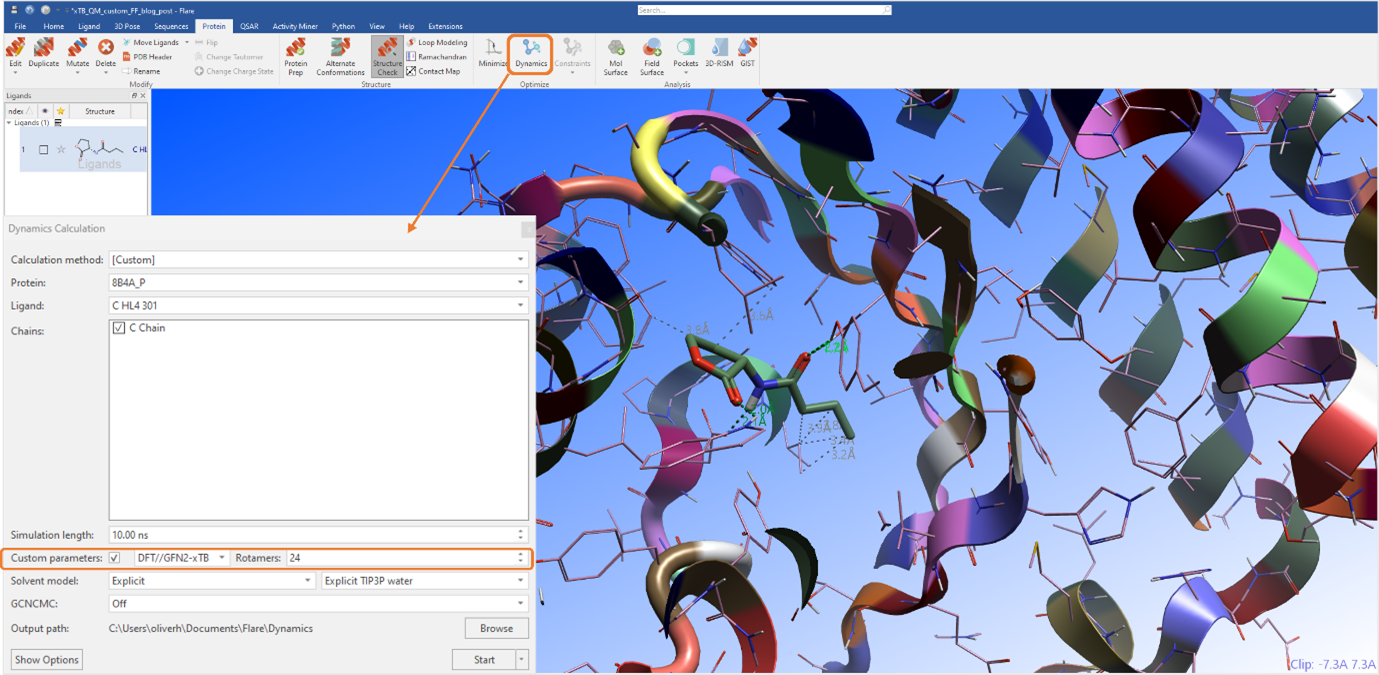
Figure 1: MD experiment with DFT//GFN2-xTB custom force field parameterization.
The custom force field parameterization calculation begins with fragmenting the molecule about each rotatable bond, while preserving the electronic environment around the bond, using Wiberg Bond Order (WBO)-based fragmentation.1 Changes in the WBO are indicative of changes in the molecular functionality around the rotatable bond of interest and so the parent (molecule) is fragmented to the smallest possible entity until the WBO of the rotatable bond of interest is no longer conserved. At this point, the parent molecule cannot be fragmented any smaller, as this would be altering the electronic environment about the rotatable bond, and the obtained fragment is carried forward into the torsion scan. This method ensures a computational speedup by at least 4-12 times, without negatively affecting the representability of the torsional potential energy profile.
Torsional potential energy scans are performed, for each fragment, at 15° intervals using wavefront propagation at semi-empirical GFN2-xTB level of theory. In the hybrid DFT//GFN2-xTB workflow, an additional B3LYP-D3BJ/DZVP single-point energy calculation is performed on each rotamer geometry to further improve the accuracy of the torsional potential energy profile. This combination of DFT functional and basis set was chosen as it balances computational efficiency against accurately reproducing the conformational energetics of neutral, positively, and negatively charged molecules calculated using higher level quantum mechanical calculations, such as Coupled Cluster (CC).2–4
A thorough validation of the hybrid DFT//GFN2-xTB approach was conducted on a large library of over 550 small molecules, encompassing a very wide range of different chemistries as well as, -2, -1, +1, and neutral charge states. For each of these small molecules, the torsional potential energy profile for each rotatable bond was calculated using a full QM torsion scan, namely, a B3LYP-D3BJ/DZVP derived torsion potential on the full parent molecule without fragmenting. Full QM torsion scans using this quantum mechanical method, as referenced above, replicate CC computed torsional potentials and, as such, offer a good approximation to the true ground-state molecular torsional potential. The validation shows that the hybrid approach computes torsional potential energy profiles within 1.08 kcal/mol relative to the reference profile computed using a full QM torsion scan.
This validation study showcased how the hybrid DFT//GFN2-xTB workflow improves the accuracy of the computed torsional potential energy surfaces relative to using GFN2-xTB alone and replicates the more expensive, full QM approach (Figure 2).
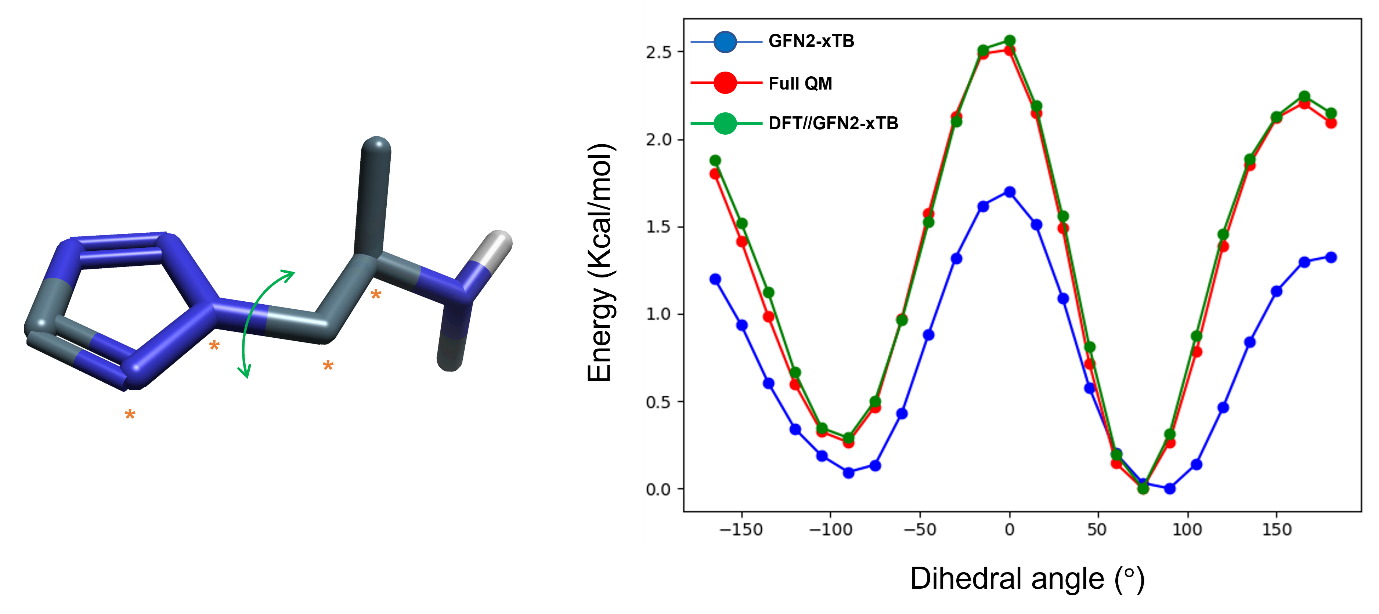 Figure 2: DFT//GFN2-xTB (green) largely improves the torsional potential energy profile relative to GFN2-xTB alone (blue) and can replicate the profile predicted at full QM level (red). For the fragment depicted (left), the asterisks denote the four atoms that form the dihedral angle being scanned at 15° intervals.
Figure 2: DFT//GFN2-xTB (green) largely improves the torsional potential energy profile relative to GFN2-xTB alone (blue) and can replicate the profile predicted at full QM level (red). For the fragment depicted (left), the asterisks denote the four atoms that form the dihedral angle being scanned at 15° intervals.
The DFT B3LYP-D3BJ/DZVP single-point energy calculation does not incur large additional computational overhead costs, only extending the GFN2-xTB custom parameterization workflow by a few minutes. This hybrid method offers a far more attractive choice to obtain accurate ligand torsional potential energy surfaces compared to executing full QM torsion scans about each rotatable bond.
In this article, we have discussed how the new hybrid DFT//GFN2-xTB workflow available in Flare can generate accurate torsional potential energy profiles for custom force field parameterization of novel molecules. This new method has the potential to improve the ability of any novel molecule to explore adequately its conformational space improving the accuracy of MD and FEP experiments.
Request a free evaluation of Flare today to further explore its full portfolio of molecular modeling capabilities.
1. Wiberg, K. B. Application of the Pople-Santry-Segal CNDO Method to the Cyclopropylcarbinyl and Cyclobutyl Cation and to Bicyclobutane. Tetrahedron 1968, 24 (3), 1083–1096. https://doi.org/10.1016/0040-4020(68)88057-3.
2. Kesharwani, M. K.; Karton, A.; Martin, J. M. L. Benchmark Ab Initio Conformational Energies for the Proteinogenic Amino Acids through Explicitly Correlated Methods. Assessment of Density Functional Methods. J. Chem. Theory Comput. 2016, 12 (1), 444–454. https://doi.org/10.1021/acs.jctc.5b01066.
3. Qiu, Y.; Smith, D. G. A.; Boothroyd, S.; Jang, H.; Hahn, D. F.; Wagner, J.; Bannan, C. C.; Gokey, T.; Lim, V. T.; Stern, C. D.; Rizzi, A.; Tjanaka, B.; Tresadern, G.; Lucas, X.; Shirts, M. R.; Gilson, M. K.; Chodera, J. D.; Bayly, C. I.; Mobley, D. L.; Wang, L.-P. Development and Benchmarking of Open Force Field v1.0.0—the Parsley Small-Molecule Force Field. J. Chem. Theory Comput. 2021, 17 (10), 6262–6280. https://doi.org/10.1021/acs.jctc.1c00571.
4. Řezáč, J.; Bím, D.; Gutten, O.; Rulíšek, L. Toward Accurate Conformational Energies of Smaller Peptides and Medium-Sized Macrocycles: MPCONF196 Benchmark Energy Data Set. J. Chem. Theory Comput. 2018, 14 (3), 1254–1266. https://doi.org/10.1021/acs.jctc.7b01074.