Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Free Energy Perturbation (FEP) methods enjoy a strong reputation for delivering reliable and robust binding affinity predictions for small molecule drugs. The accuracy of prediction is typically very high – to within 1 kcal/mol of the experimental value. This has exciting ramifications for drug discovery projects, where reliable affinity predictions are required before committing to the synthesis and testing of new potential leads.
One barrier to using FEP can be time constraints and computational resources which scale with the number of molecules in the dataset. These issues can be mitigated by taking advantage of a workflow starting with a fast and high- throughput method of R-group exploration in order to cast the net wide, followed by refining and filtering for fewer molecules with the most potential. Using FEP in this way is efficient and enables one to make the right decisions for the best molecules to make and test with greater confidence. An example of such a workflow will be investigated and discussed.
This case study references the structure-based optimization campaign demonstrated by Hoon Han et al. J. Med. Chem, 2020 for the discovery of noncovalent inhibitors targeting the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2 3CLpro). The particular test for the following workflow was to examine if the highest affinity binder found in Table 1 of this reference (‘Compound 21’) could be re-discovered, by simulating a drug discovery workflow starting from little information (a couple of known actives and a PDB reference code 7LME).
The case study presented here demonstrates a fast and efficient workflow, see Figure 1, minimizing the risks of wasting time on ‘bad’ molecules and also of missing the ‘good’ ones.
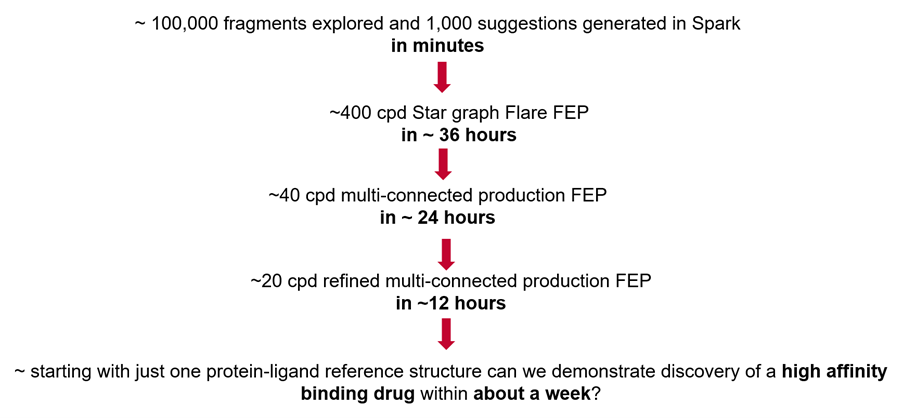
Figure 1. The Spark ⇒ Flare FEP workflow used in this case study simulates the discovery of a non-covalent high affinity SARS-CoV-2 inhibitor in approximately a week of work.
Spark is the leading market bioisostere replacement solution to generate innovative ideas in your discovery project by exploring biologically relevant replacements (bioisosteres) for key fragments of known active compounds.
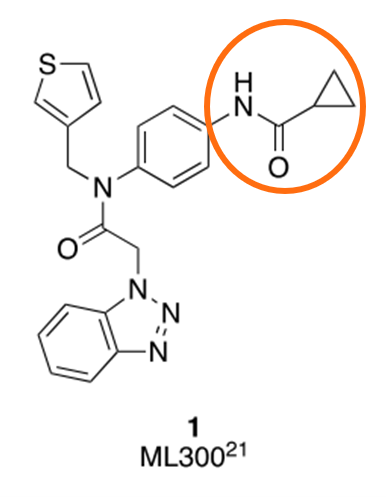
Figure 2. Known active 'ML300' from PDB 7LME, the co-crystal starting point used by Hoon Han et al. J. Med. Chem, 2020 and in this case study. The R-group replacement region selected for bioisostere replacement in Spark is circled in orange.
In this example we conduct a simple Spark experiment starting from PDB 7LME. The co-crystal ligand ‘ML300’ is chosen as the ‘reference’ molecule for drug development through R-group modification (Figure 2), and the protein to act as a volume exclusion. The region of ML300 to explore in R-group replacement is circled in Figure 2. The Spark R-group exploration is run by searching through ~97,000 fragments from the ‘CHEMBL_Common’ and ‘Very Common’ and ‘Common’ commercial Spark databases.
In the second part of this workflow, Flare FEP was used to triage the Spark results and choose the most promising molecules to progress based on a stepwise workflow with increasing accuracy of binding affinity predictions.
Using Star graph mode to generate your perturbation network in FEP is an efficient first phase for an initial triage of many molecules. Figure 3 compares 3 Star graphs i) of 12 compounds ii) of 250 compounds and iii) of ~400 compounds (250 + automatically generated intermediates), all linked to a center known active. Example i) clearly shows that there is only 1 link between the center molecule and each design possibility (each R-group modification). If intermediates are added to aid the more difficult transitions then the link number can increase, but compared to fully connected perturbation graphs (as in ‘Normal multi-connected production mode:’, below) the number of links is much more limited. Star graph networks offer a good balance between a slightly lower precision (due to less connectivity and lack of cycle error analysis) and much higher speed. They enable the user to quickly triage a very large number of compounds with reasonable accuracy, from which the top highest predictions can be taken forward to the next step of FEP calculations with improved accuracy.

Figure 3. Comparing a i) 12 compound Star graph ii) 250 compound Star graph with no intermediate generation and iii) ~400 compound Star graph with automatic intermediate generation.
From the Spark experiment, the top 250 of 1000 hits were taken forward sorted by %BIF (which represents the percentage of similarity to the starter molecule, ML300). A fast 1ns λ window simulation (single way) was ran using ‘Compound 9’ (a known active from Table 1 in Hoon Han et al. J. Med. Chem, 2020) as the center of the Star or ‘parent’ molecule. The R-group modified candidates from Spark (the edges on the Star graph) by design share the same scaffolds with 'Compound 9', and in any case where the transformation is difficult (the ligands are very different), intermediate automation implemented in Flare FEP will improve the success of the perturbation.
After a couple of Star graph runs the top 40 compounds with highest affinity were then taken forward into a normal multi-connected production mode run, again with 'Compound 9' used as the ‘known active’. A longer 4ns λ window simulation (dual way) was used (versus the 1ns λ window single way simulations in the fast Star graph runs). The ‘normal’ mode gives increased precision due to more connectivity and cycle error analysis, whilst the longer sampling time also typically improves the quality of the calculation combined with dual way transformations that provide information on any significant hysteresis effects.
The most promising 16 ligands (top binders with the most trusted results) from the 40 compound run where then taken forward to a second multi-connected production mode run. Ligands with higher uncertainty of prediction were discarded and more links and intermediate structures were added to strengthen the remaining cycles. Furthermore for some of the links the number of λ windows was increased to achieve better overlap, thereby refining and honing in on the details.
At this point, 'Compound 21' stands out as the best predicted binder (highest affinity) from the initial ~100,000 fragment wide search.
The final Flare FEP run in this workflow was performed on a reduced number of ligands in the perturbation graph to focus on the those with most potential, and using a longer sampling time (10 ns). This final run predicts the binding affinity for 'Compound 21' at -8.9 +/- 0.8 kcal/mol, in good agreement to the -9.8 kcal/mol experimental affinity.
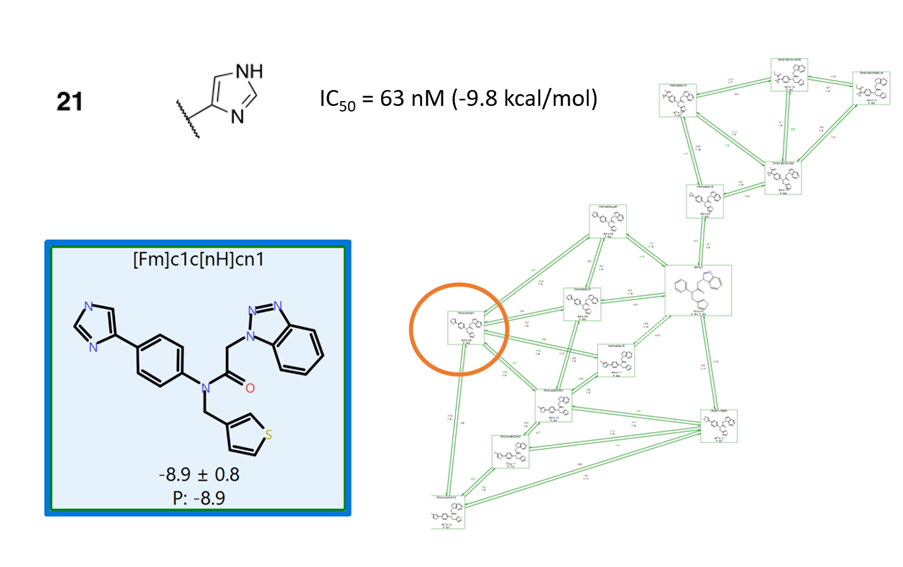
Figure 4. 'Compound 21' is discovered in this workflow with a predicted binding affinity of -8.9 +/- 0.8 kcal/mol (compared to -9.8 kcal/mol measured affinity)
In this case study, the discovery of ‘Compound 21’ was in fact efficiently and easily reproduced using the workflow proposed in Figure 1. Here the discovery of 'Compound 21' is the main focus, but moreover this process opens up other options to pursue (for example the top cycle in Figure 4 contains alternative arrangements of the ML300 cyclopropyl moiety which are predicted to be high affinity binders). The workflow uses Spark to explore widely the chemical space, selecting the most promising R-groups from a start point of ~100,000 options. Flare FEP is then used to triage results down to a handful of potentials with accurately predicted binding affinities, leading to the identification of some of the ‘best molecules’ for your drug discovery project within a very reasonable time-frame.
See the flexible licensing options for Flare and Spark, and request an evaluation to try them on your project.
Register to attend the webinar ‘Using Flare FEP to prioritize the best molecules in drug discovery’ on December 7th.