The adaptability of pyFlare to access advanced data visualization and calculation functionalities
We showcase some examples within a drug discovery context where the pyFlare environment has been further expanded with advanced data ...
News
For any computational chemist, the ability to run several different calculations in parallel can significantly increase the efficiency of their drug design workflow. In previous versions of Flare™, each calculation was required to end before another experiment could begin. Thanks to recent updates to the platform, which offers a full portfolio of ligand-based and structure-based methods, users are now able to perform multiple experiments simultaneously, within a single Flare project. With this new multiprocessing ability, Flare features a redesigned 'Calculations’ window that collectively monitors all jobs after they have been submitted. As an example, Figure 1 shows a docking calculation, a Molecular Dynamics (MD) simulation, and a Quantum Mechanics (QM) calculation all running together in parallel, alongside a protein preparation job which has recently been completed.
For each experiment, the calculations window reports errors for troubleshooting. The window also shows the progress of current calculations, the computing resources being used, and a log file of terminated jobs. These details, accessible within one window, enable the user to manage various experiments more effectively.
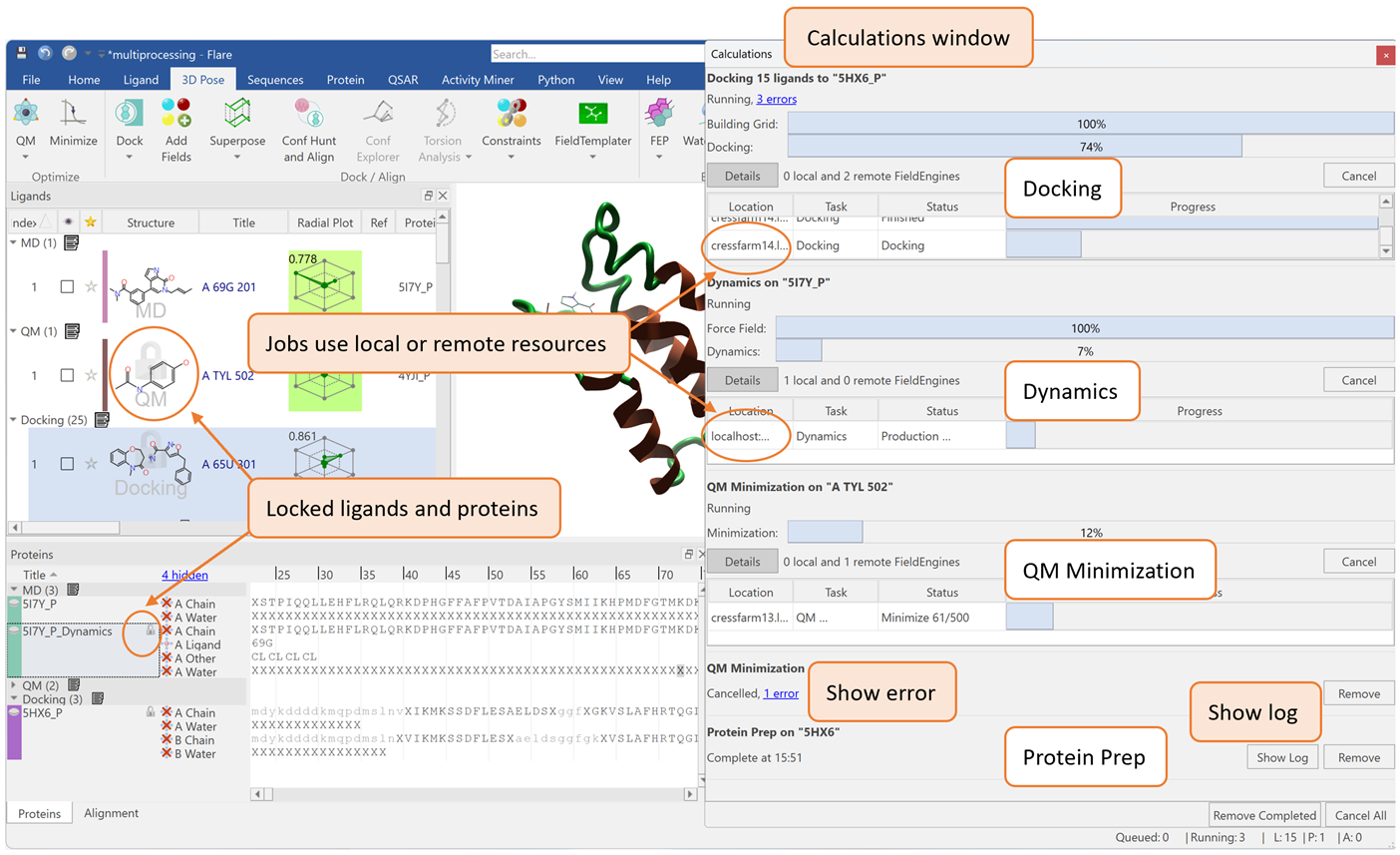
Figure 1. Multiple experiments running together in parallel within Flare.
In the context of multiprocessing, Flare users can allocate specific computing resources to individual experiments. For this, Flare’s enhanced 'Start’ button (Figure 2) allows chemists to choose whether a calculation should run locally or remotely, via Cresset Engine Broker™ (CEB), a Linux® application designed to speed up computational chemistry calculations by running them on a remote server or cluster.
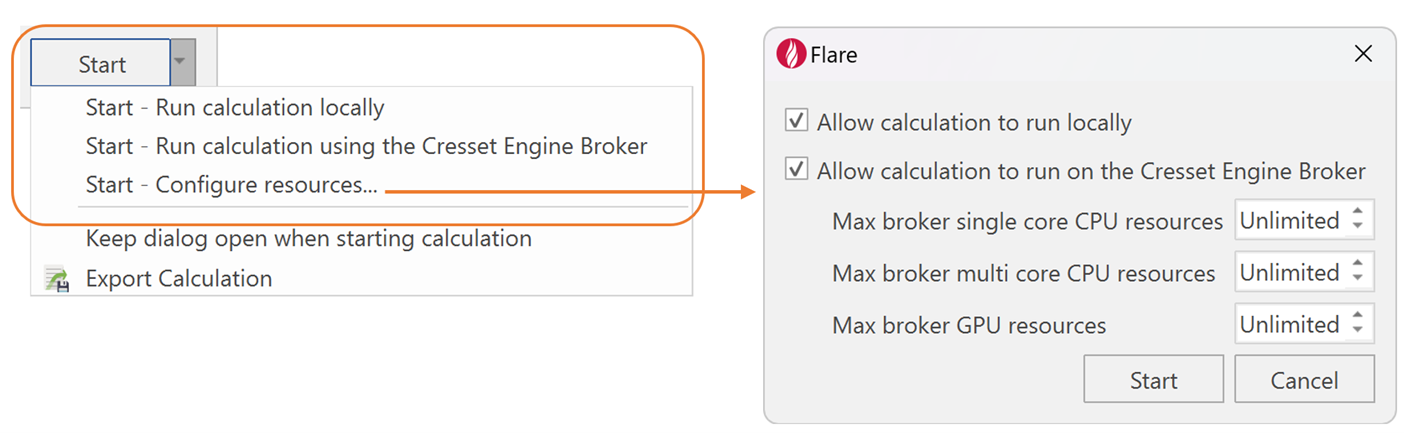
Figure 2. Allocating available computing resources to a chosen experiment within Flare.
For example, the user may apply all local resources for one given experiment (e.g., MD), while simultaneously starting other jobs (e.g., docking and QM) using the CEB (Figure 1). If instead, the user submits jobs without choosing any of those options, Flare will automatically determine the most efficient use of both local and remote resources.
To ensure that parallel experiments run seamlessly, Flare locks ligands and proteins of an active calculation so that these molecules cannot be modified by any other concurrent operations. This intelligent locking prevents accidental editing of input structures, preserving the integrity of each experiment. In the GUI, locked ligands and proteins will have a lock symbol, as shown in Figure 1.
Learn how Flare’s multiprocessing capabilities could bring efficiencies and advancements to your drug discovery workflow – request an evaluation today.