Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
We are delighted to announce the release of Spark V10.3, Cresset’s idea and bioisostere generator. This release of Spark brings many enhancements, including:
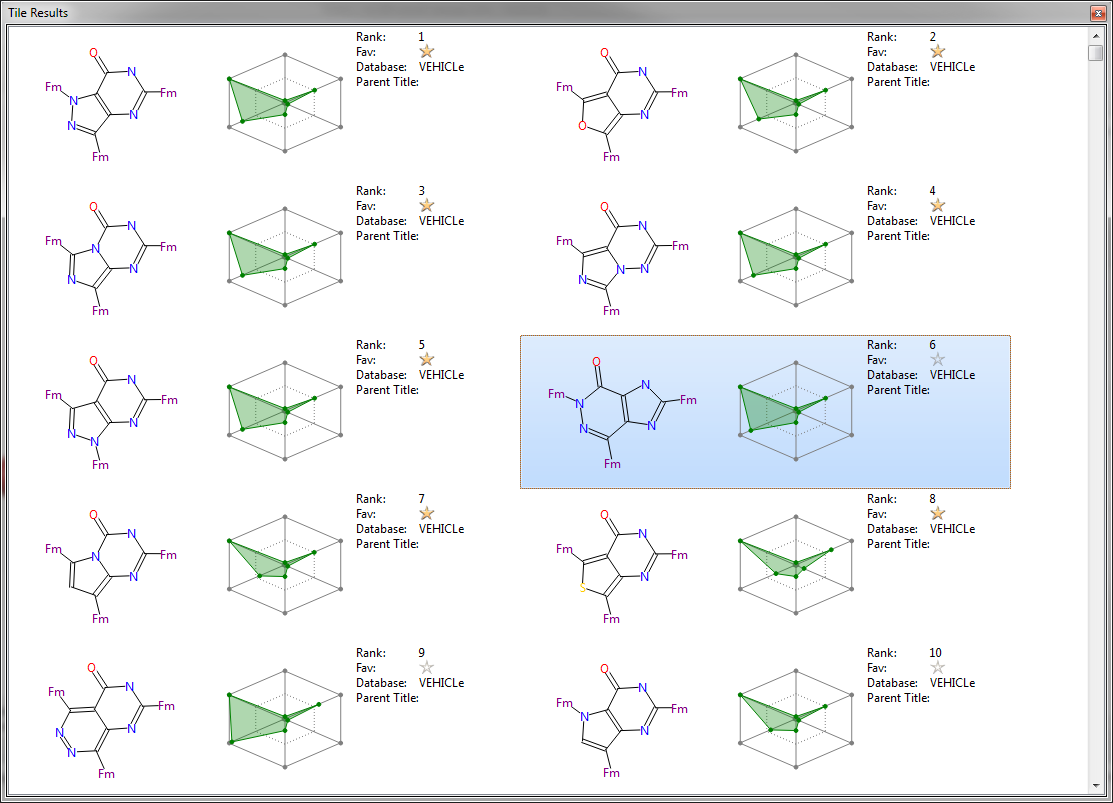
This feature was leaked in the sneak peek in the September newsletter (together with the new ‘tags’ feature) but it’s worth highlighting it again. Each result molecule is presented on a tile which can be configured to hold exactly the data that you want to see. We like this view so much that we now use it as our default and have it configured with a mixture of structure, physical properties (shown in the radial plot) and selected textual data, such as the database that was used. The final outcome is a rapid way to scan through the results to find those which you want to follow up.
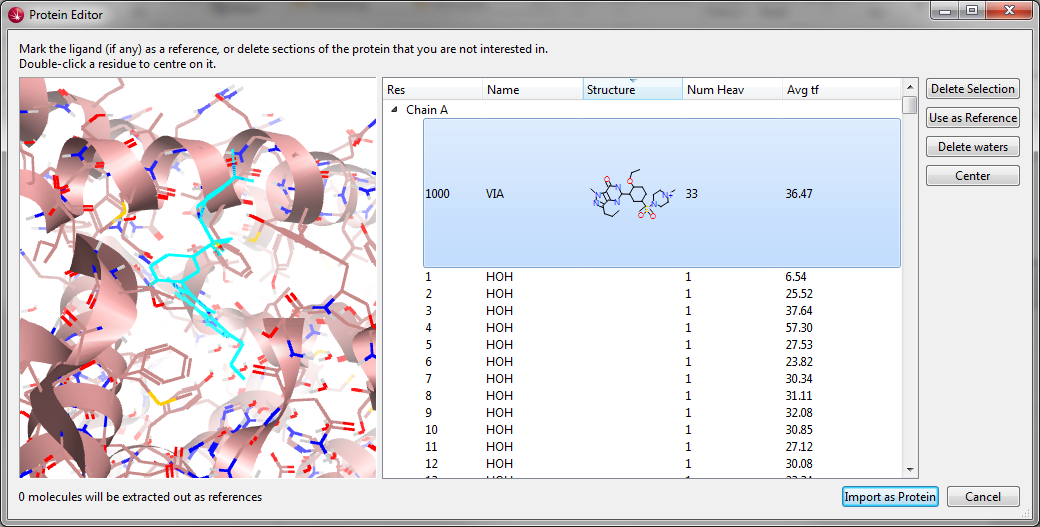
The current release of Forge and Torch includes a simple parser for pdb files that has now been introduced into Spark. Using the new protein import wizard you are able to load a pdb file, quickly identify the ligand, remove water molecules and import the protein. When combined with the powerful molecular editor it is now easy to split ligand-protein crystal structures into ligand and protein, edit the ligand to ensure correct bonding and geometry, run a search for new cores or R-groups for the ligand and display the results in the protein active site.
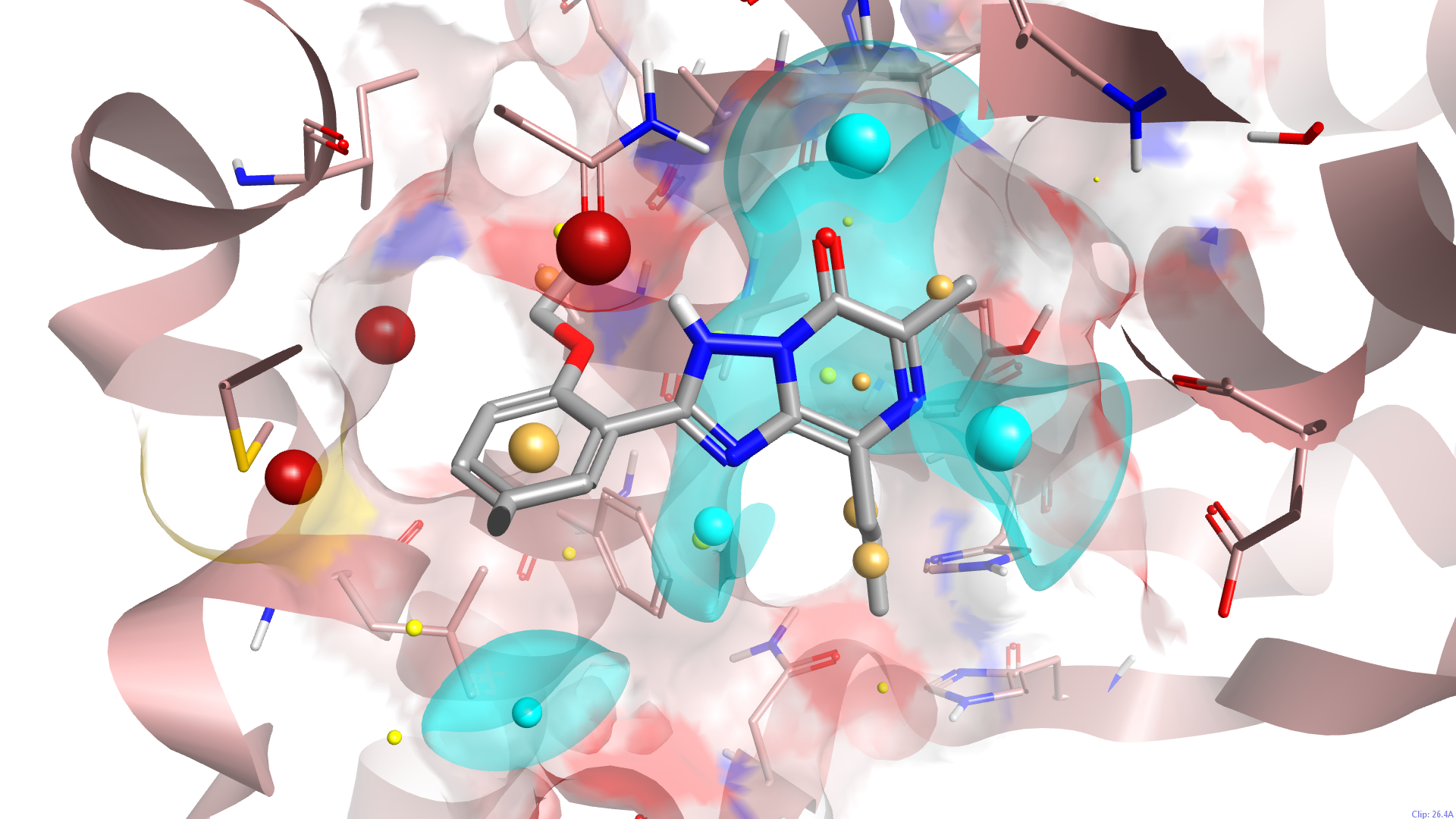
As well as improving the import of proteins this release of Spark brings enhanced visualization options for proteins and ligand-protein interactions. A new button toggles the display of protein residues to those immediately around your ligand. A separate button gives control over the protein backbone display and new options for protein-ligand interactions enables the display of potential hydrogen bonds or protein clashes. The result is significantly improved graphical handling of proteins.
Spark has always assessed new bioisosteres in the context of the final molecule, i.e. we work in product space. We think that this is the only sensible place to score our potential replacements as it enables both electronic and steric influence to be calculated between the retained and introduced portions of the molecule. To do this every result molecule is energy minimized so that replacements are scored in an energetically reasonable conformation wherever possible. This release introduces greater control over this minimization and also over the minimization of fragment conformations that are stored in the database. Now you can choose the exact extent to which molecules are relaxed before scoring so that you see more of the conformation effects that can dominate some cases (like the replacement of flexible chains).