Improving PROTAC properties via single-point changes to linkers
We explore how computational methods can be applied to proteolysis targeting chimera (PROTAC) design, to effectively tackle some of the ...
News
Computational chemistry is a dynamic field, characterized by an abundance of tools with new ones being added every year. Free Energy Perturbation (FEP) calculations have emerged as an essential component of this arsenal, since they help researchers to accurately predict the binding affinity of molecules, significantly contributing to the design and prioritization of new drugs. Recognizing this immense potential of FEP at Cresset, we have developed Flare™ FEP – a powerful, innovative solution designed to make FEP calculations accurate, efficient, and easy to run. Our vision extends beyond the release of another product; we aim to democratize the power of FEP, so it is considered not a privilege for a few, but a standard method available to all. With Flare FEP, we envisage a future where anyone, regardless of their technical background, can take advantage of the power of free energy calculations, applying them across diverse projects to unlock new possibilities and make the molecules that matter.
Flare FEP's innovative features have been designed with this universal accessibility in mind. They can be used from the beginning to the end of your project to streamline your FEP calculations and include tools for preparing the molecular system of interest, run and troubleshoot the FEP calculation, analyze the results and decide the next steps. Adaptive lambda scheduling, automatic generation of intermediates, and custom parameter derivation are only a few examples, and they allow anyone, no matter their expertise level, to bring the power of FEP calculations into their project. In this article, these unique features will be explored in more detail.
To better guide you through the unique technical features of Flare FEP, let’s suppose that we want to run a short FEP calculation, using a set of congeneric ligands with known experimental activities. The first thing that the user will notice when opening a Flare project is its user-friendly interface, an element that is common to all Cresset software solutions. The design of Flare strikes a balance between technical sophistication and usability. The protein of interest is loaded and prepared in Flare, as well as the ligands, which are aligned to the co-crystalized ligand of the target using our patented ligand alignment method to quickly generate an excellent starting point for the FEP calculations. Within minutes, the system that will be used for the FEP calculation is ready.
The next step involves the generation of the FEP graph, that connects the molecules of the congeneric series to define the alchemical transformation. Flare FEP's usability and user-friendliness is enhanced by the automated graph generation using LOMAP and the capability to predict and incorporate intermediate molecules into the FEP graph.1 In instances where a transformation is too complex to occur in a single step, the software intelligently identifies these situations and inserts intermediate molecules into the FEP graph. This not only enhances the accuracy of the simulation but also smoothens the pathway from the initial to the final state, ensuring that the transformations are computationally feasible. This level of automation, guided by the smart use of LOMAP, minimizes user intervention, ensuring a more streamlined, efficient, and accurate FEP simulation. The afore-mentioned steps that initiate the FEP experiment can be completed within minutes as shown in Figure 1.
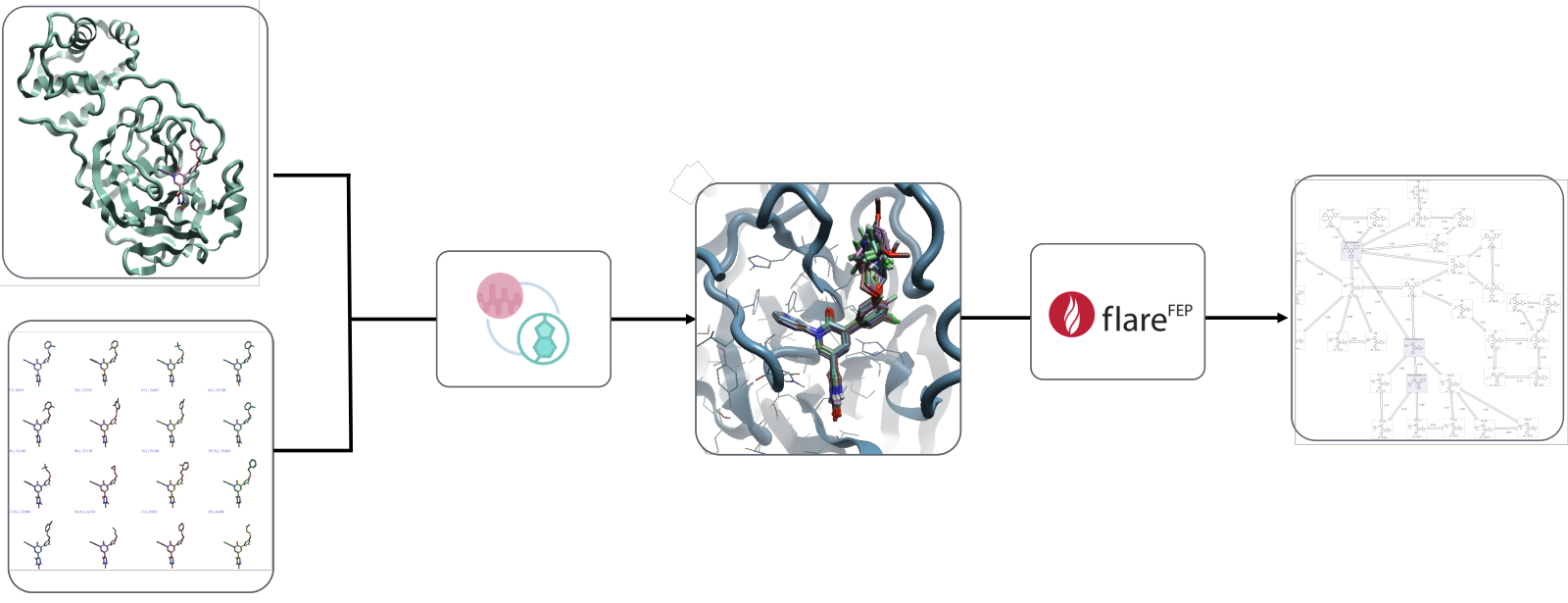
Figure 1: Alignment of ligands using the Maximum Common Substructure (MCS). Import into the FEP project and creation of the FEP graph can be completed in a few minutes. The workflow starts with a crystal structure of a protein and a set of ligands.
Having generated the FEP graph, the calculation itself needs to be run. Each transformation consists of many, independent Molecular Dynamics (MD) simulations that happen across a λ coordinate that ensures a smooth transition between the different states. Running MD simulations means that the ligands and the protein should be parameterized with a forcefield: Flare offers both the Amber and OpenFF forcefields for the parameterization of small molecules.2-4 The latter allows derivation of custom forcefield parameters for the torsions of small molecules, a capability that Flare FEP leverages to deliver highly accurate results. By allowing for customized, high-quality torsion parameters, the software can cater to the unique structural and dynamic characteristics of each ligand. This approach may enhance the precision of the simulations and ensure a more accurate representation of the molecule's behavior. Users are thus able to rely on a more refined forcefield model that is tailor-made for their specific ligands, significantly enhancing the accuracy of their FEP calculations on some systems.
The quality of predicted free energies depends on the efficient sampling of the events within the binding pocket. For this reason, modeling solvent effects accurately is essential. Proper sampling allows for the assessment of solvent contributions, including desolvation and reorganization, which can significantly impact free energy calculations. Advanced sampling techniques are often necessary for accurate predictions and for this reason, Flare FEP is equipped with a Grand Canonical Non-Equilibrium Candidate Monte Carlo (GCNCMC) method that rehydrates the binding site by inserting and deleting water molecules within a sphere around the ligand in the binding site (Figure 2).5 Unlike normal Grand Canonical Monte Carlo methods that use Metropolis acceptance criteria, the non-equilibrium candidate sampling technique improves the probability of a successful water insertion/ deletion, making the algorithm more efficient.
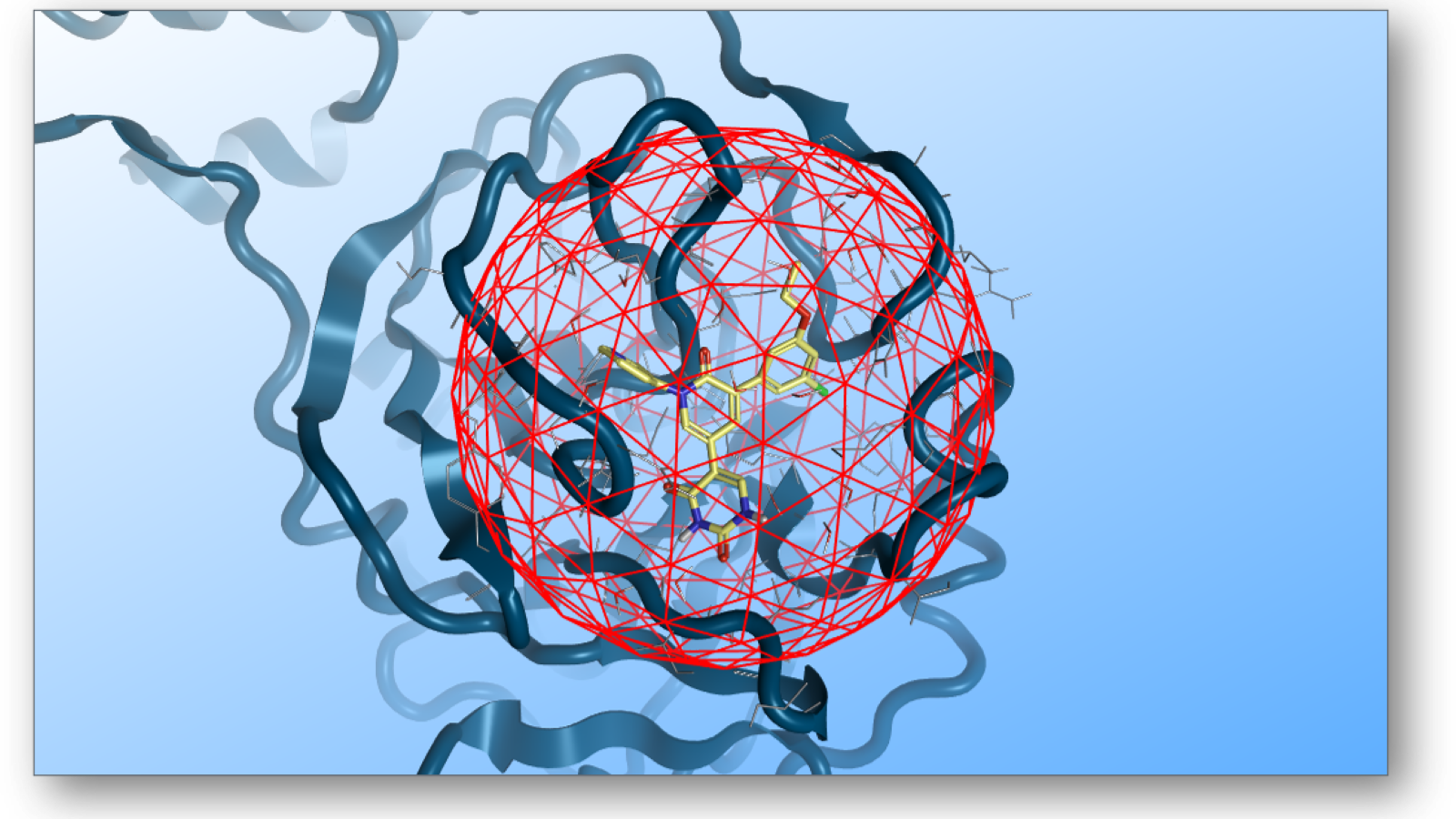
Figure 2: GCNCMC inserts and deletes water molecules in a sphere around the ligand, capturing efficiently solvation effects in the binding site. The radius of the sphere can be defined by the user.
One of the standout features of Flare FEP that significantly contributes to its speed and reduces the computational cost is the ‘adaptive lambda window algorithm’ which is summarized in Figure 3. Traditionally, the choice of lambda values for ligand transformations has been static and often arbitrary, leading on some occasions to inefficient sampling and consequently inaccurate results. On the contrary, Flare FEP’s unique adaptive lambda window algorithm runs very short MD calculations in the free phase (solely ligand in solvent) and calculates the overlap between the frames of the lambda states before the actual FEP calculation, determining the optimal number of lambda windows necessary to ensure convergence of the FEP calculations. This adaptive nature balances perfectly accuracy and computational efficiency. Instead of using a fixed, potentially excessive number of lambda windows, which can lead to unnecessary simulations and increased costs, Flare FEP's adaptive approach ensures that only the minimum required windows are used. This effectively optimizes the time and cost involved in running FEP calculations. Thus, reliable results can be obtained in a shorter time frame and at a reduced cost.
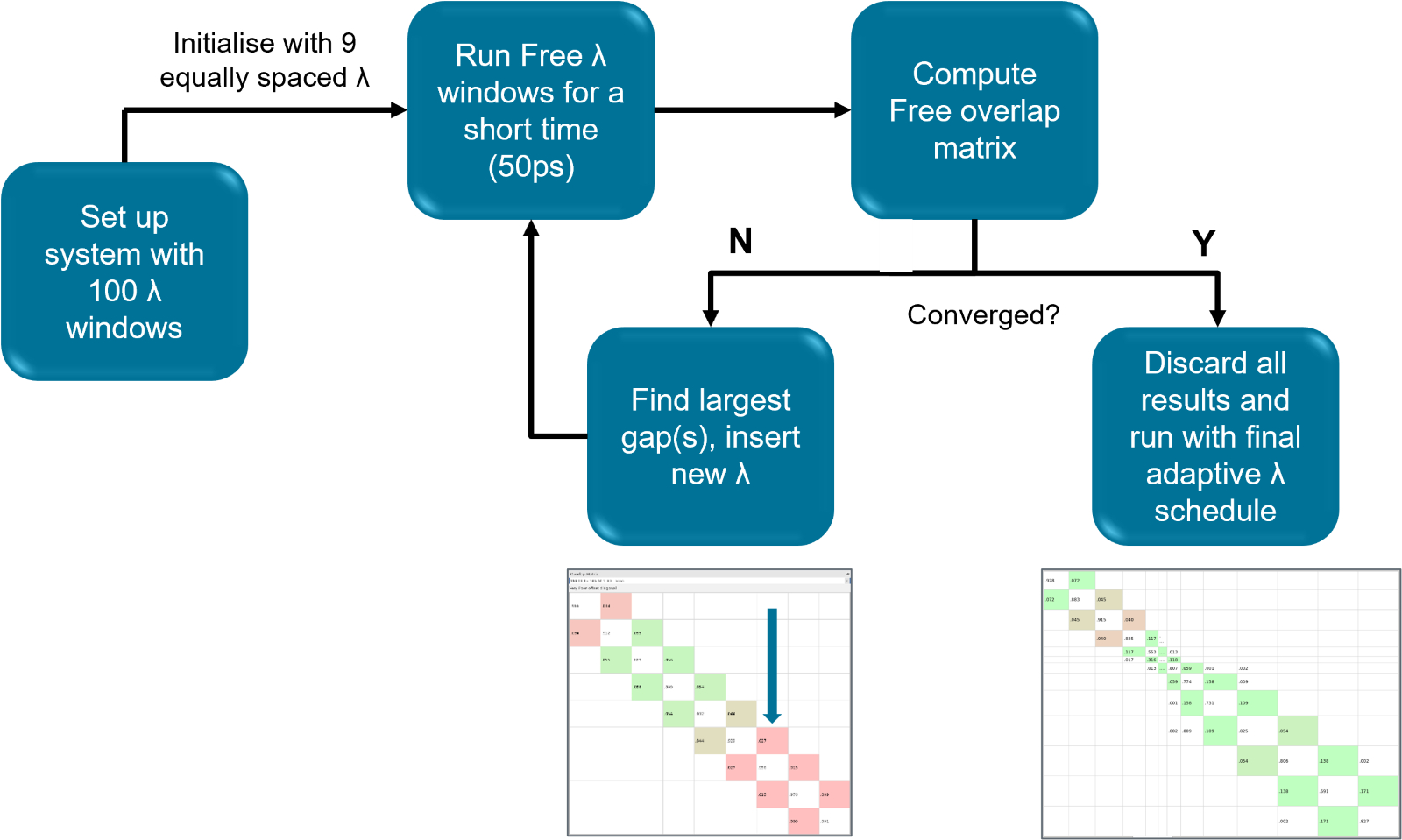
Figure 3: The adaptive lambda algorithm ensures that the minimum number of lambda windows is used for each transformation, keeping the computational cost to the minimum without affecting the accuracy of the calculation.
Flare FEP's usability is further improved with its GPU-accelerated technology, which allows FEP calculations to be run in a wide spectrum of hardware configurations, spanning from latest technology to consumer-grade Graphical Processing Units (GPUs). This capability enables researchers to use easily available and cost-effective hardware for their complex computational chemistry tasks. With the assistance of the Cresset Engine Broker™, an application that bridges client computers (running on Windows, Linux, or macOS) with high-performance computing resources such as a Linux cluster, researchers can harness the power of GPUs for their FEP calculations. This factor makes it an attractive choice for computational chemistry teams working within a tight budget but still seeking top-tier software. Moreover, for those requiring extensive computational resources or seeking a cloud-based solution, Flare FEP also supports running calculations via Amazon Web Services (AWS). This flexibility in hardware and cloud options provides an inclusive platform suitable for a range of computational demands, highlighting Flare FEP's commitment to user-centric design and functionality.
Getting back to our example, as soon as the benchmarking FEP calculation is finished we want to analyze the results to validate the model. However, like all computational methods, FEP calculations are not free from potential pitfalls and sources of error. Incorporating a range of troubleshooting tools within the software offers several advantages.
The first tool that is used after running a calculation is the ‘activity plot’ (Figure 4, right), that shows the experimental versus the predicted activity values, so the user can assess the accuracy of the model using statistics such as R2 and the Mean Unsigned Error (MUE). In Flare FEP the activity plot is equipped with ‘sub-graph analysis’ (Figure 4, left), a feature that helps in identifying subsets of molecules with lower internal error statistics (if any).
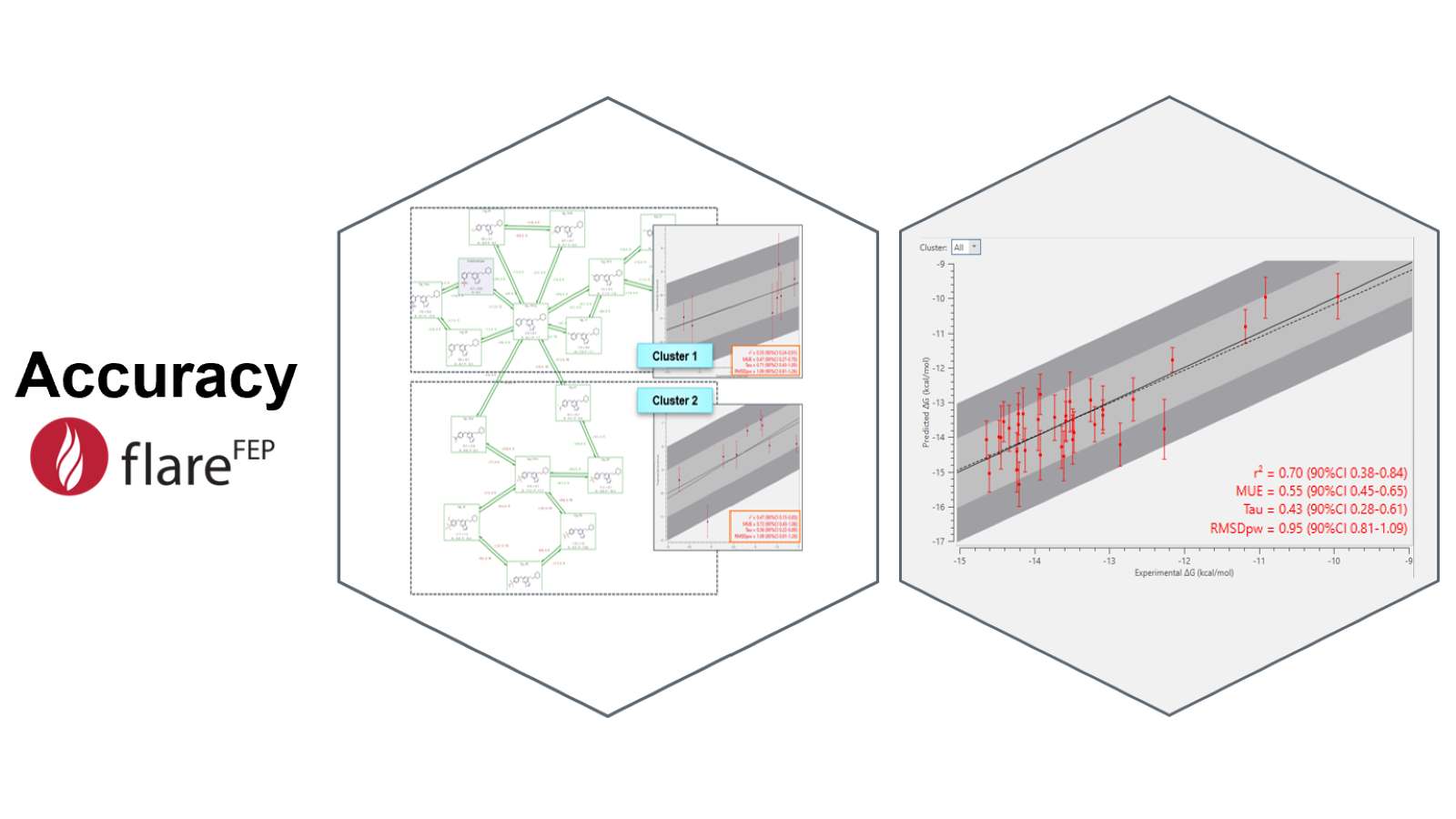
Figure 4: The activity plot and the sub-graph analysis are two tools of Flare FEP that help you assess the accuracy of your FEP experiment.
When it comes to issues with sampling and convergence, Flare FEP offers a range of tools that help with the identification of possible issues. ‘Link plots’ (Figure 5, left), which display the calculation of free energy differences (ΔG) using techniques like Thermodynamic Integration (TI) and the Multistate Bennett Acceptance Ratio (MBAR), along with contributions from each lambda window, play a vital role in identifying sampling issues during free energy calculations. By comparing results from TI and MBAR, we can gain insights into the consistency and convergence of the FEP calculation. Examining the contributions from each lambda window highlights regions of the simulation where sampling might be inadequate or problematic. Discrepancies between TI and MBAR results or unusual patterns in the contributions may signal issues like insufficient sampling, hysteresis, or slow conformational transitions.
Cycle error and hysteresis analysis offer a way to spot inconsistencies and potential errors in the calculations. ‘Cycle errors’ (Figure 5, center) can reveal discrepancies when comparing the free energy obtained from a direct path versus its reverse. ‘Hysteresis’, on the other hand, indicates if a system hasn't reached equilibrium, which might lead to inaccurate free energy calculations.
Plotting the ‘overlap matrices’ (Figure 5, right) of the transformations is also crucial for assessing the quality of sampling between neighboring states in a simulation, as poor overlap can lead to unreliable free energy differences. Flare FEP offers tools that visually highlight areas of poor overlap and helps researchers make more informed decisions on refining the simulation parameters (e.g., change the options for a specific link).
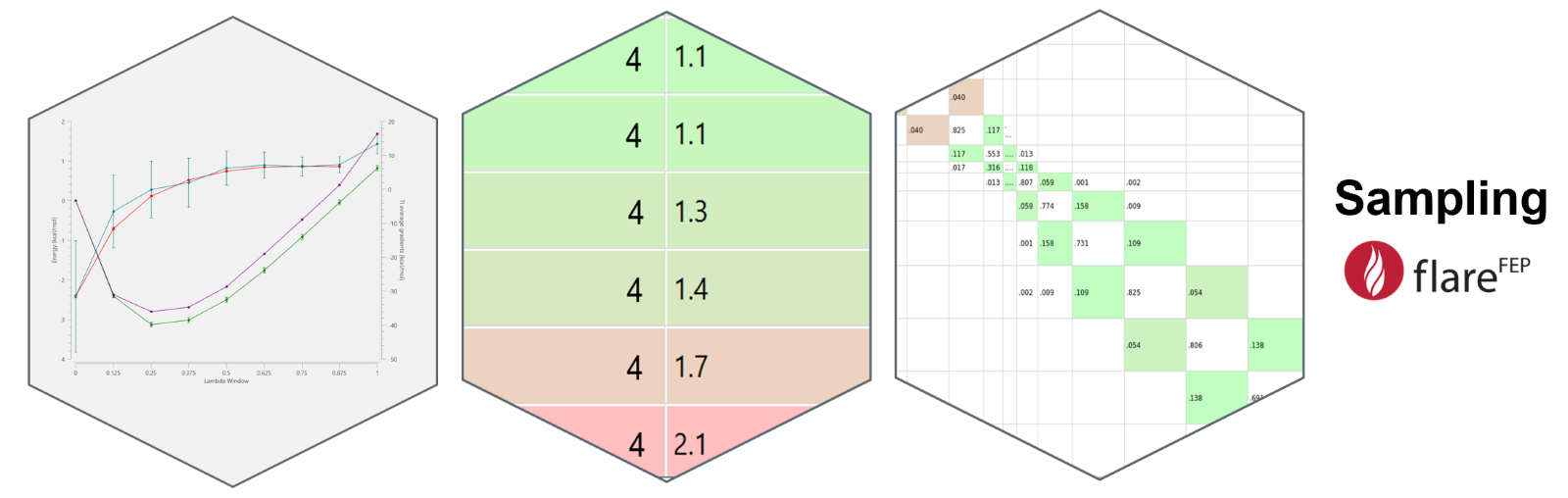
Figure 5: Link plots, cycle errors and the overlap matrix allow us to identify possible sampling issues that may lead to inconsistencies at the calculations.
Visualization is key in molecular simulations. The ‘3D window’ in Flare FEP allows users to inspect the conformation of the transformed ligand and ensure that the ligand doesn't adopt any unrealistic or undesired geometries during the simulation, which could bias results. The 3D structure of the ligand is shown as a snapshot of the final frame of the trajectory for the beginning (λ= 0) and at the end of the transformation (λ= 1) for both the forward and the backward transformation. The 3D window is also equipped with tools that are available in Flare, such as visualization of molecular surfaces as shown in Figure 6, left. Moreover, the FEP graph (Figure 6, right) not only shows the transformations of the ligands, but it also provides useful information about the calculation. The experimental (if available) and calculated binding affinities are shown beneath the 2D representation of the ligand. The relative difference in free energy (ΔΔG) is shown above the links, which are also color-coded to highlight any hysteresis.
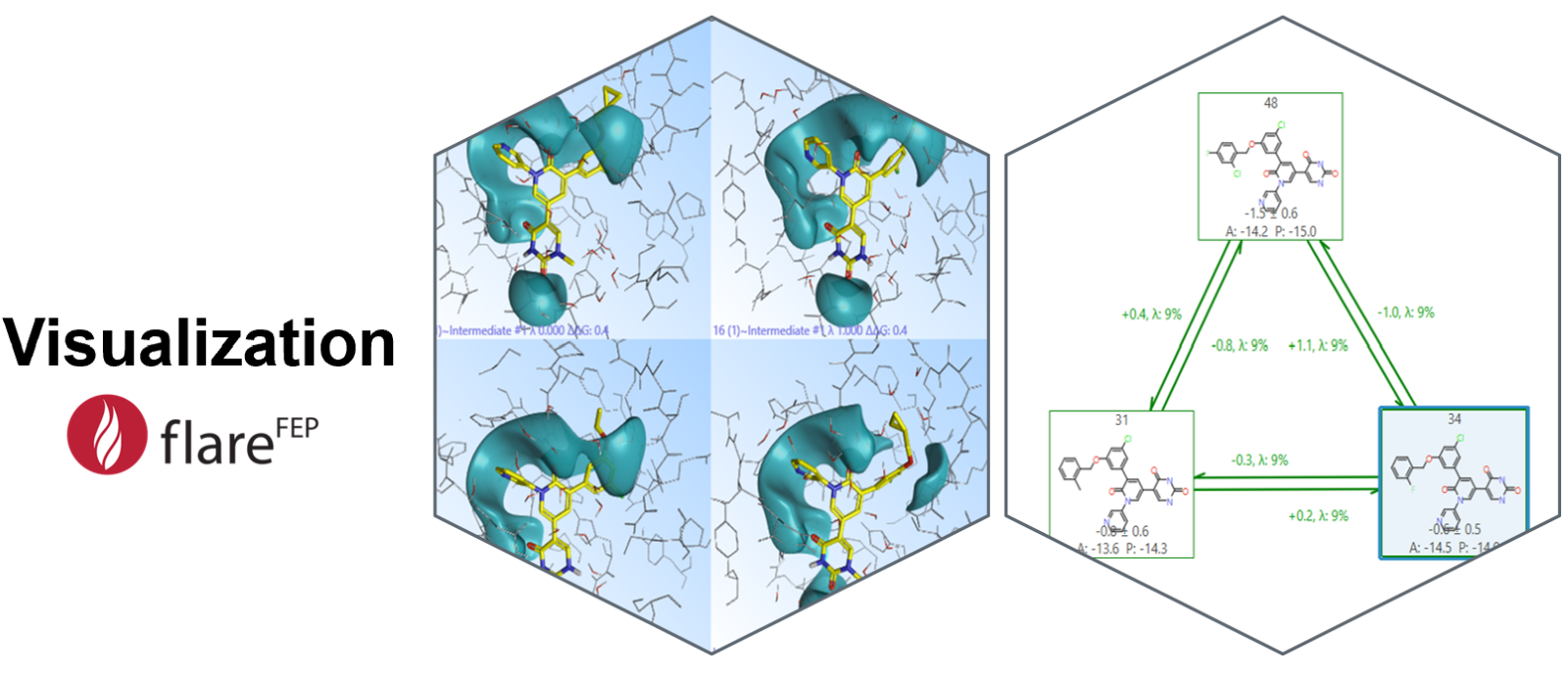
Figure 6: Visualisation tools in Flare FEP allow inspection of the initial and final states of the molecules in 3D, as well as inspection of the transformations through the FEP graph.
The usability of Flare FEP is not limited to features used to run the benchmark FEP experiment, but offers tools that simplify and accelerate the process of generating new design ideas by means of integrated workflows that combine Flare FEP with other tools and products, such as Hit Expander or Spark™.
Hit Expander's strength lies in its ability to effortlessly generate numerous simple molecular variants by substituting atoms within a molecule's structure. The specific positions for substitution can be selected, and then atoms or R-groups are inserted in every amenable place, generating new designs in seconds.
Spark, Cresset’s bioisosteric replacement and scaffold hopping tool, allows fast exploration of diverse chemical space. By suggesting replacements for specific molecular fragments, Spark enables researchers to think creatively about chemical structure modifications. This tool leverages on a vast database of potential bioisosteric replacement fragments derived from real molecules to suggest alternatives that maintain or enhance the desired biological activity.
Both Hit Expander and Spark can be seamlessly integrated into the Flare FEP workflows, aiming at the generation of novel molecules with improved pharmacological profiles. Design ideas generated by Hit Expander and Spark can be taken to the next level with the ‘Quick FEP’ option in Flare FEP. This feature enables users to perform rapid free energy perturbation calculations on the designed molecules, quantitatively assessing the potential impact of structural modifications on binding affinities or other relevant properties. By doing so, we can efficiently triage and prioritize the most promising candidates for further investigation.
Some recent case studies exemplify the potential of these integrated workflows in Flare FEP. In the first case study, our goal was to assess whether an in silico workflow could facilitate the discovery of active CDK9 inhibitors—a crucial target in cancer research. Firstly, we ran a Hit Expander experiment in conjunction with Quick FEP. In doing so we retrieved highly active molecules and made sensible suggestions for further modifications to improve activity. Through a swift computational screening process with Hit Expander, the SAR landscape around a specific molecule was explored and 84 new ligands were generated. These molecules were then triaged with ‘Quick FEP’ using a ‘minimum spanning tree’ graph. The best 20% of these molecules were then refined using a second FEP experiment, with a normal, multi-connected graph and several promising candidates with remarkable binding affinities to the CDK9 target were identified. What is worth mentioning is that this entire process, from ideation to identification of potential drug candidates, was completed in less than 24 GPU hours.
The second case study, summarized on this poster, demonstrates the effectiveness of Flare FEP calculations, when used in conjunction with Spark, in guiding strategic chemical modifications to enhance the potency and selectivity of potential inhibitors of SARS-Cov-2 Mpro cystein protease. Through benchmarking studies, a correlation was established between experimental and predicted binding affinities (R2 = 0.55, MUE = 0.58 kcal/mol) for a set of inhibitors identified in previous research. Using Spark, new molecular designs were generated, and they were evaluated with Flare FEP. Notably, among the designs from Spark, at least 13 molecules exhibited activity equal to or greater than the original compound.
Figure 7: The new molecular designs that were generated from Spark are imported into Flare and aligned on the co-crystalized ligand, to start the FEP experiment.
These collaborative approaches, combining Flare FEP with other software solutions and modules, offer a powerful toolkit for designing and evaluating novel molecules. Hit Expander and Spark, when coupled with the speed and accuracy of Flare FEP not only hold promise in accelerating the discovery of novel therapeutics, but they also empower researchers to explore uncharted territories of chemical space.
Having explored the unique technical features of Flare FEP and having seen two examples of its usability, it becomes clear that Cresset’s solution stands out in this competitive landscape. Ultimately, the technical features translate into faster, more accurate results, and substantial cost savings. Flare FEP continues to exhibit notable performance enhancements as we introduce fresh features and methodologies. This is visually represented in the graphs presented below, which showcase a comparison of our software's accuracy in relation to previous versions (depicted in various shades of blue) and the results of Wang et al. (illustrated in purple).6 The final set of bars in the graphs provides an average assessment across all the molecular systems investigated. Remarkably, the R2 values achieved, on average, surpass our previous performance, and the MUE is approximately 0.10 kcal/mol lower than earlier iterations of Flare. It is noteworthy that the accuracy of Flare FEP is comparable to that reported by Wang et al.6 for these specific molecular systems.
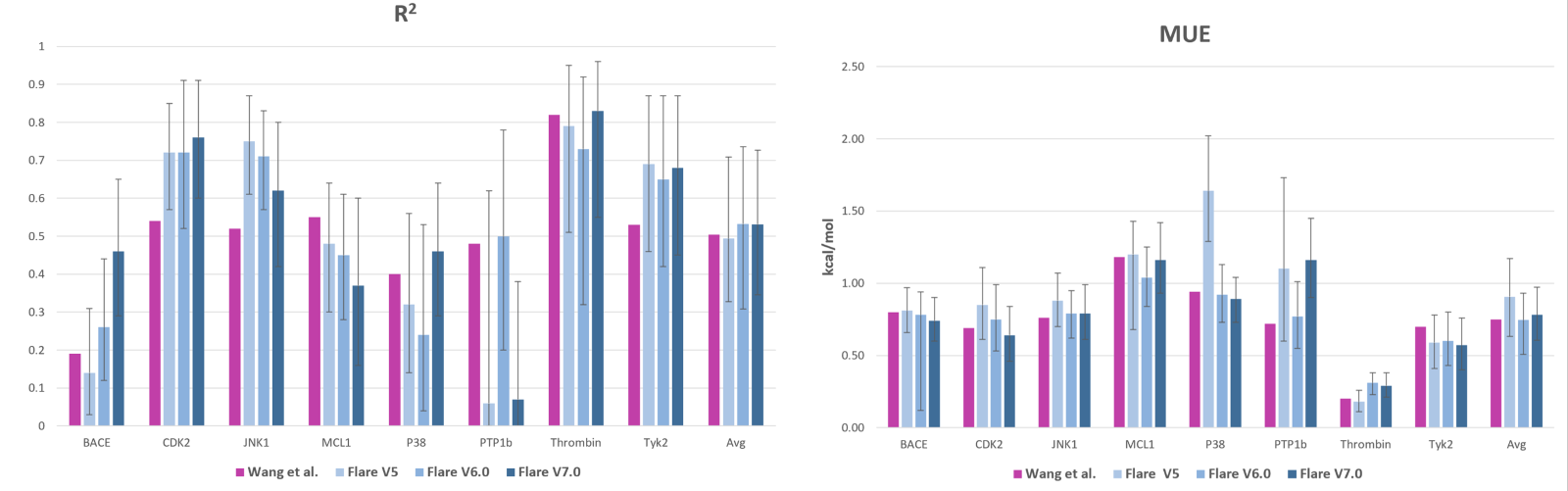
Figure 8: Comparison of MUE and R2 between the three latest releases of Flare and the original calculations that were reported by Wang et al.6
However, Flare FEP is appealing to users not only because of its features but also thanks to its affordability. While other FEP tools in the market often come with a big price tag that may discourage many research groups and academic institutions, Flare FEP offers competitive pricing.
In conclusion, Flare FEP brings an innovative approach to free energy calculations in computational chemistry. Its technical prowess, coupled with a user-friendly interface, is set to redefine how computational chemists approach their work, providing a tool that's not only efficient but also cost-effective. Whether you are an established computational chemist, or a student still finding their way within the world of computational chemistry software, Flare FEP offers a comprehensive, intuitive, and powerful tool that can help streamline your workflows and enhance your research outcomes. Our message is clear and uncompromising: FEP should not be a privilege, but a standard!
Make the right ligand design choices and enable lead optimization with confidence – request an evaluation and try Flare FEP on your project today. During evaluation, you’ll have the chance to put Flare FEP’s full range of unique features to the test, while having the freedom to publish any results produced and use these for further research.